One of the most common posttranslational modifications of eukaryotic proteins is glycosylation. Glycosylation of proteins can affect many biological activities. For therapeutic glycoproteins, it can modify biological activity, targeting, trafficking, serum half life, clearance, and recognition by receptors (1, 2). For such reasons, biomanufacturers must monitor and characterize the glycosylation patterns of their recombinant therapeutic proteins (3, 4).
Therapeutic proteins have two main types of glycosylation: N-linked glycans and O-linked glycans (5). Attachment of an N-glycan starts in the endoplasmic reticulum (ER), where a core nascent glycan is linked through the side-chain amide nitrogen on specific asparagine residues of a protein at sites that have the sequence NXS/T, where X can be any amino acid residue except proline. N-glycans are trimmed and further modified as a glycoprotein transits through the ER and Golgi apparatus. Host cell and cell culture conditions can alter the type of N-glycosylation present on the glycoprotein (from high mannose to complex and hybrid N-glycans). O-glycosylation occurs in the Golgi apparatus. N-glycans have a common core consisting of two N-acetylglucosamine (GlcNAc) residues and three mannose (Man) residues. But the only common core for O-glycans is the N-acetyl-galactosamine (GalNAc) residue attached through the oxygen atom on the side chain of serine or threonine residues of a protein.
Glycosidases: Glycosylation is complex and heterogeneic, so multiple analytical methods must be used to determine the structure of glycans and their location on a glycoprotein. Glycosidases are important tools often used with other analytical methods to remove, trim, or modify glycans. Glycosidases (glycoside hydrolases) are enzymes that hydrolyze the glycosidic bonds of complex sugars. Such enzymes are applied in three main analytical areas for therapeutic glycoproteins: removing glycans for analysis, trimming glycans for sequencing, and modifying glycans in glycoengineering. Herein we describe the different enzymes used in each area with specific examples of their applications.
Removal of Glycans for Analysis
The most commonly used enzyme for the removal of N-glycans from a glycoprotein is peptide-N-glycosidase F (PNGase F). It is an amidase that cleaves between the innermost GlcNAc residue of the N-glycan and the asparagine residue that releases the N-glycan. That creates an aspartic acid residue in place of the asparagine residue on the protein.
One main reason that glycosidases are so frequently used is that they have a very broad specificity cleaving high mannose, complex, and hybrid N-glycans. The only limitation for such enzymes is that one cannot cleave if there is an α1–3 fucose linked to the core GlcNAc residue (the sugar residue next to the protein). Such modification occurs only in plant, insects, mollusks, and parasitic worms. N-glycans harboring this modification can be removed by the enzyme PNGase A.
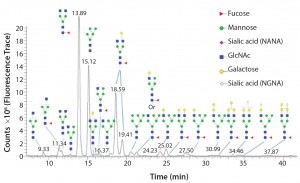
Figure 1: Glycoforms identified by LC/MS analysis of intact Erbitux (cetuximab) digested with PNGase F
Once an N-glycan has been released by an amidase, it can be fluorescently labeled for analysis by liquid chromatography coupled to fluorescence detection (LC–FLD) and/or mass spectrometry (LC–MS) or capillary electrophoresis coupled to laser induced fluorescence detection (CE–LIF) analysis. Figure 1 illustrates the variety of N-glycans released from the Erbitux therapeutic antibody (cetuximab) by PNGase F and detected by LC–MS analysis.
In addition to PNGase F, many different endo-β-N-acetylglucosaminidases also release N-glycans from glycoproteins. Most of those endoglycosidases are limited in the type of N-glycans that they can recognize and cleave. Table 1 shows the different specificities of most commercially available endo-b-N-acetylglucosaminidases. All of those endoglycosidases hydrolyze the N,N′diacetyochitobiose moiety (between the two GlcNAc residues on the N-glycan core) leaving a GlcNAc residue attached to the protein. So they often are used to determine occupancy or presence of an N-glycan at a specific site. That is particularly useful for glycoproteins with multiple N-glycosylation sites.
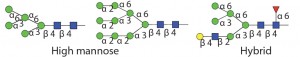
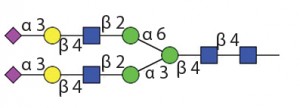
Table 1 Endo H and Endo F1: Substrate specificity of commercially available endoglycosidases ([purple diamond] sialic acid, [yellow circle] galactose, [green circle] mannose, [royal blue square] GlcNAc, [red triangle] fucose)(14)
![Table 1 Endo H and Endo F1: Substrate specificity of commercially available endoglycosidases ([purple diamond] sialic acid, [yellow circle] galactose, [green circle] mannose, [royal blue square] GlcNAc, [red triangle] fucose)](https://lne.e92.mwp.accessdomain.com/wp-content/uploads/2016/02/Table2-300x68.jpg)
Table 1 Endo F2: Substrate specificity of commercially available endoglycosidases ([purple diamond] sialic acid, [yellow circle] galactose, [green circle] mannose, [royal blue square] GlcNAc, [red triangle] fucose)(15)
![Table 1 Endo H and Endo F1: Substrate specificity of commercially available endoglycosidases ([purple diamond] sialic acid, [yellow circle] galactose, [green circle] mannose, [royal blue square] GlcNAc, [red triangle] fucose)](https://lne.e92.mwp.accessdomain.com/wp-content/uploads/2016/02/Table3-300x79.jpg)
Table 1 Endo F3: Substrate specificity of commercially available endoglycosidases ([purple diamond] sialic acid, [yellow circle] galactose, [green circle] mannose, [royal blue square] GlcNAc, [red triangle] fucose)(15)
![Table 1 Endo H and Endo F1: Substrate specificity of commercially available endoglycosidases ([purple diamond] sialic acid, [yellow circle] galactose, [green circle] mannose, [royal blue square] GlcNAc, [red triangle] fucose)](https://lne.e92.mwp.accessdomain.com/wp-content/uploads/2016/02/Table4-300x83.jpg)
Table 1 Endo S: Substrate specificity of commercially available endoglycosidases ([purple diamond] sialic acid, [yellow circle] galactose, [green circle] mannose, [royal blue square] GlcNAc, [red triangle] fucose)(16)
![Table 1 Endo H and Endo F1: Substrate specificity of commercially available endoglycosidases ([purple diamond] sialic acid, [yellow circle] galactose, [green circle] mannose, [royal blue square] GlcNAc, [red triangle] fucose)](https://lne.e92.mwp.accessdomain.com/wp-content/uploads/2016/02/Table5-300x66.jpg)
Table 1 Endo S2: Substrate specificity of commercially available endoglycosidases ([purple diamond] sialic acid, [yellow circle] galactose, [green circle] mannose, [royal blue square] GlcNAc, [red triangle] fucose)(17)
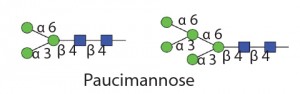
Table 1 Endo D: Substrate specificity of commercially available endoglycosidases ([purple diamond] sialic acid, [yellow circle] galactose, [green circle] mannose, [royal blue square] GlcNAc, [red triangle] fucose)(18)
![Table 1 Endo M: Can also do transglycosylation reactions. Substrate specificity of commercially available endoglycosidases ([purple diamond] sialic acid, [yellow circle] galactose, [green circle] mannose, [royal blue square] GlcNAc, [red triangle] fucose)(19)](https://lne.e92.mwp.accessdomain.com/wp-content/uploads/2016/02/Table7-300x143.jpg)
Table 1 Endo M: Can also do transglycosylation reactions.
Substrate specificity of commercially available endoglycosidases ([purple diamond] sialic acid, [yellow circle] galactose, [green circle] mannose, [royal blue square] GlcNAc, [red triangle] fucose)(19)
Currently, no known broad specificity endoglycosidase can cleave all O-glycans from glycoproteins. So analysis of O-glycans is far more difficult than for N-glycans. Two O-glycosidases are commercially available. One is from Streptococcus pneumoniae and can release Core 1 disaccharides consisting of a galactose (Gal) linked β1–3 to N-acetylgalactosamine (GalNAc), where the GalNAc residue is alpha linked to either a serine or threonine residue of the protein (7). The other O-glycosidase is from Enterococcus faecalis and has a slightly broader specificity, cleaving both the Core 1 disaccharide and the Core 3 disaccharide that consists of GlcNAc linked β1–3 to GalNAc (8).
Neither enzyme can cleave if the disaccharide is further modified by sialic acids or other sugars. Intact O-glycans can be released chemically using b-elimination, but care must be taken with this method not to degrade the released glycans.
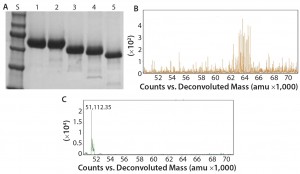
Figure 2: Deglycosylation of the etanercept therapeutic protein, a fusion protein with three N-glycans and up to 13 O-glycans: (A) SDS-PAGE gel of etanercept treated with different combinations of PNGase F and exoglycosidases; lane S is the molecular weight standards. Lane 1 is the untreated etanercept control. Lane 2 is etanercept incubated in the presence of buffer containing SDS and DTT. Lane 3 is etanercept incubated with PNGase F in the presence of a buffer containing SDS and DTT. Lane 4 is etanercept incubated with a deglycosylation mix, which is a mixture of PNGase F, O-glycosidase (from Enterococcus faecalis), an α2–3,6,8 neuraminidase, a β1–4 galactosidase, and a broad specificity β N-acetylglucosaminidase in the presence of SDS and DTT. Lane 5 is etanercept incubated with Rapid PNGase F, O-glycosidase, and an α2–3,6,8 neuraminidase in a proprietary Rapid PNGase F buffer. Complete removal of all glycans indicated by a complete shift of etanercept only occurs with conditions in lane 5. (B) ESI-TOF profile of etanercept before treatment (average molecular weight = 64 KDa); (C) after treatment (51 kDa) with Rapid PNGase F, O-glycosidase and an α2–3,6,8 neuraminidase in a proprietary Rapid PNGase F buffer for 1 hour at 37 °C, demonstrating that all N- and O-glycans have been removed.
Another major disadvantage to this method is that the protein is destroyed and can no longer be analyzed for its structure or activity. By contrast with intact chemical release of O-glycans, the O-glycans can be trimmed using a combination of exoglycosidases and an O-glycosidase (Figure 2). Although this method can preserve a protein’s structure and activity, it degrades the glycans so they cannot be characterized. In addition, some O-glycans contain sugar modifications (e.g., sulfation or acetylation) that render modified sugars resistant to cleavage by exoglycosidases.
Trimming Glycans for Sequencing
Exoglycosidases are important enzymes that can trim glycans one sugar residue at a time from the nonreducing end of a glycan. A large number of different exoglycosidases are commercially available.
Exoglycosidases are specific for both the type of sugar and its anomer (alpha or beta). Some exoglycosidases are more general and can hydrolyze a number of different linkages. For example, a broad-specificity neuraminidase can remove sialic acid residues linked α2–3, α2–6, α2–8, or α2–9 to a glycan. Other exoglycosidases are more specific for a particular linkage. For example, a β1–4 galactosidase will remove only β1–4 linked galactose. Because of the innate specificity of these enzymes, exoglycosidases are useful for both trimming glycans and determining the sequence of the glycans. Basic LC-MS or matrix-assisted laser desorption/ionization (MALDI) analysis can determine only the size of a glycan. However, to confirm the linkage or type of monosaccharide present on a glycan, it is necessary to perform tandem mass spectrometry (MS-MS), such as collision-induced dissociation (CID) or use exoglycosidases followed by CE or MS analysis.
When using exoglycosidases, it is important to consider the quality of enzyme preparation. Enzyme preparations isolated from native sources often can be contaminated with other glycosidase activities, making glycan structure elucidation more difficult. Recombinant sequencing-grade versions of most commonly used exoglycosidases are now commercially available for use in sensitive glycan analysis workflows.
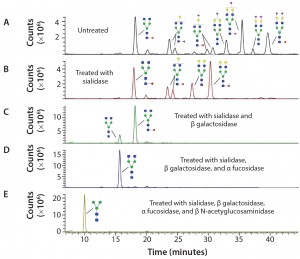
Figure 3: Sequencing of Enbrel glycans; the glycans have been released from Enbrel using PNGase F, labeled with procainamide, treated with different glycosidases and analyzed using LC–MS; (A) LC trace of glycans release from Enbrel with no exoglycosidase treatment; (B) LC trace of release glycans treated with α2–3,6,8,9 Neuraminidase A; (C) LC trace of release glycans treated with α2–3,6,8,9 neuraminidase A and β1–4 galactosidase S; (D) LC trace of release glycans treated with α2–3,6,8,9 neuraminidase A, β1–4 galactosidase S and α1–2,4,6 fucosidase; (E) LC trace of release glycans treated with α2–3,6,8,9 neuraminidase A, β1–4 galactosidase S, α1–2,4,6 fucosidase and β-N-acetylglucosaminidase S. Glycan structures were assigned to specific major peaks deduced from the molecular weight of the labeled glycans and confirmed by exoglycosidase digestion.
Figure 3 shows how exoglycosidases can sequence glycans released from the Enbrel therapeutic protein (etanercept). Exoglycosidases also can be used to sequence glycans still attached to a glycoprotein. These enzymes are especially useful for analyzing and detecting potential antigenic structures on glycans. For example, using a general alpha galactosidase such as the enzyme from green coffee bean to treat therapeutic proteins can help identify low levels of Galα1–3Gal (α Gal epitopes) that are not present in humans and can be immunogenic (9).
Modification of Glycans
In addition to removal of intact glycans and glycan sequencing, glycosidases can be used to modify glycans on glycoproteins (termed glycoengineering) (10). One method is to remove the undesired N-linked glycans using endoglycosidases. Then the desired uniform glycans created using chemoenzymatic synthesis can be attached to the glycoprotein through the GlcNAc residue left by the endoglycosidase cleavage. Attachment of the desired glycan can be performed using endoglycosidases (which naturally transglycosylate) such as Endo-M (11) or using endoglycosidase mutants that drive the reaction toward addition of a glycan instead of removal of a glycan (12) (Figure 4).
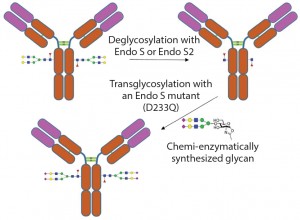
Figure 4: Example of enzymatic transglycosylation for glycoengineering of an antibody to create homogenous glycoforms
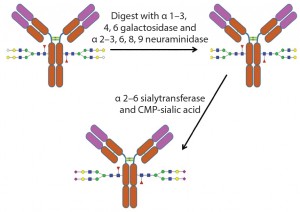
Figure 5: Example of the use of exoglycosidases and a sialyltransferase to create uniform glycosylation on an antibody
An alternative method of glycoengineering involves using exoglycosidases to trim glycans down to a uniform size on a glycoprotein. Such glycans can then be reconstructed using specific glycosyltransfereases, enzymes that transfer monosaccharides from a nucleotide sugar donor to a glycan. That creates a glycoprotein with a more homogenous glycan structure (Figure 5).
Future Applications
So far, these in vitro glycoengineering methods have been used mostly at small scales. The exception is for glycoprotein enzymes used to treat lysosomal storage diseases. Several of these enzymes have been glycoengineered using exoglycosidases to expose mannose structures on a protein to improve the enzyme’s transport to the lysosome by receptor-mediated endocytosis (13). In addition, modification of glycosylation pathways in host cells (including Roche’s GlycArt and Kiran’s Potelligent expression systems) are being used to produce therapeutic proteins with desired glycosylations. Therapeutic protein production in those cell lines is well under way, with several therapeutic proteins in clinical trials.
Scientists expect that the speed and simplicity of glycan analysis will progress with further improvements in enzymes and in analytics as the need for higher-throughput methods increases. A released formulation of PNGase F called Rapid PNGase F by New England Biolabs and GlycoWorks Rapid PNGase F by Waters has drastically improved the speed of N-glycan removal from antibodies and many other therapeutic proteins from overnight to five minutes.
New antibody-specific proteases such as IdeS and IdeZ are improvements over typical trypsin/LysC or papain digestions of antibodies because they specifically cleave at the hinge region of IgG with no secondary site cleavage. Use of those proteases with improved software allows MS analysis of the constant region of an antibody. That allows determination of the types of glycans present on the Fc without the need to remove, purify, and label N-glycans.
Other areas of improvement might lead to the discovery of new enzyme specificities. For example, an enzyme with a broad specificity for complex O-glycans would allow better analysis of these types of protein modification. And a PNGase that could remove all N-glycans would be very useful as more therapeutic proteins are expressed in transgenic plants and insect cell lines. Finally, as methodologies for glycan analysis become more refined with higher throughput and increased sensitivities, it should be possible to select for clones during development or use selected glycoengineered expression hosts that produce antibodies with desired glycosylation patterns. Such speed and sensitivity will also allow for constant monitoring of fermentation processes, which will improve production of therapeutic glycoproteins.
References
1 Ohtsubo K, Marth JD. Glycosylation in Cellular Mechanisms of Health and Disease. Cell 126(5) 2006: 855–867; doi: 10.1016/j.cell.2006.08.019.
2 Walsh G. Posttranslational Modifications of Protein Biopharmaceuticals. Drug Discovery Today 15(17–18), 2010: 773– 780; doi: 10.1016/j.drudis.2010.06.009.
3 Beck A, et al. Trends in Glycosylation, Glycoanalysis, and Glycoengineering of Therapeutic Antibodies and Fc-Fusion Proteins. Curr. Pharm. Biotechnol. 9(6), 2008: 482–501.
4 Sethuraman N, Stadheim TA. Challenges in Therapeutic Glycoprotein Production. Curr. Op. Biotech. 17(4) 2006: 341–346; doi: 10.1016/j.copbio.2006.06.010.
5 Spiro RG. Protein Glycosylation: Nature, Distribution, Enzymatic Formation, and Disease Implications of Glycopeptide Bonds. Glycobiology 12(4) 2002: 43R–56R.
6 Wang L, et al. Structural Analysis of a Highly Glycosylated and Unliganded gp120Based Antigen Using Mass Spectrometry. Biochem. 49, 2010: 9032–9045; doi: 10.1021/bi1011332.
7 Fujita K, et al. Identification and Molecular Cloning of a Novel Glycoside Hydrolase Family of Core 1 Type O-Glycan-Specific Endo-Alpha-N-Acetylgalactosaminidase from Bifidobacterium longum. J. Biol. Chem. 280(45) 2005: 37415–37422; doi: 10.1074/jbc.M506874200.
8 Koutsioulis D, Landry D, Guthrie EP. Novel Endo-N-Acetylgalactosaminidases with Broader Substrate Specificity. Glycobiol. 18(10) 2008:799–805; doi: 10.1093/glycob/cwn069.
9 Bosques CJ, et al. Chinese Hamster Ovary Cells Can Produce Galactose-α-1,3Galactose Antigens on Proteins. Nat. Biotechnol. 28(11) 2011: 1153–1156; doi: 10.1038/nbt1110-1153.
10 Rich JR, Withers SG. Emerging Methods for the Production of Homogeneous Human Glycoproteins. Nat. Chem. Biol. 5(4) 2009: 206–215; doi: 10.1038/nchembio.148.
11 Yamamoto K, et al. Transglycosylation Activity of Mucor hiemalis Endo-Beta-N-Acetyl-Glucosaminidase Which Transfers Complex Oligosaccharides to the N-acetylglucosamine Moieties of Peptides. Biochem. Biophys. Res. Comm. 203(1) 1994: 244–252.
12 Huang W, et al. Chemoenzymatic Glycoengineering of Intact IgG Antibodies for Gain of Functions. J. Am. Chem. Soc. 134(29) 2012: 12308–12318; doi: 10.1021/ja3051266.
13 Solá RJ, Griebenow K. Glycosylation of Therapeutic Proteins. BioDrugs 24(1) 2010:
9–21; doi: 10.2165/11530550-00000000000000.
14 Trimble RB, Tarentino AL. Identification of Distinct Endoglycosidase (Endo) Activities in Flavobacterium meningosepticum: Endo F1, Endo F2, and Endo F3. Endo F1 and Endo H Hydrolyze Only High Mannose and Hybrid Glycans. J. Biol. Chem. 266(3) 1991: 1646–1651.
15 Tarentino AL, et al. Multiple Endoglycosidase F Activities Expressed By Flavobacterium meningosepticum Endoglycosidases F2 and F3: Molecular Cloning, Primary Sequence, and Enzyme Expression. J. Biol. Chem. 268(13) 1993: 9702–9708.
16 Collin M, Olsén A. Endo S: A Novel Secreted Protein from Streptococcus pyogenes with Endoglycosidase Activity on Human IgG. EMBO J. 20(12) 2001: 3046–3055.
17 Sjögren J, et al. EndoS 2 Is a Unique and Conserved Enzyme of Serotype M49 Group A Streptococcus That Hydrolyses N-Linked Glycans on IgG and α1-Acid Glycoprotein. Biochem. J. 455(1) 2013: 107– 118; doi: 10.1093/emboj/20.12.3046.
18 Mizuochi T, Amano J, Kobata A. New Evidence to the Substrate Specificity of EndoBeta-N-Acetlylglucosiamindase D. J. Biochem. 95(4) 2008: 1209–1213.
19 Yamamoto K, et al. Novel Specificities of Mucor hiemalis Endo-Beta-N-Acetylglucosaminidase Acting Complex Asparagine-Linked Oligosaccharides. Biosci. Biotechnol. Biochem. 58(1) 1994: 72–77.
Ellen P. Guthrie is a senior scientist and Paula E. Magnelli is a research scientist II at New England Biolabs, Inc., 240 County Road, Ipswich, MA 01938; 1‐978‐927‐5054; guthrie@neb.com.