In traditional sequential chromatography, columns are run as separate entities. The process requires significant hands-on time and constant manual intervention. By contrast, automated chromatography technology provides the same results more efficiently and reliably and frees researchers to focus on other tasks, thereby shortening protein purification times from days to hours.
For drug discovery, purifying protein samples is required to generate enough materials for research experiments. But the process is complex and time consuming. It involves repeated single-column purifications, careful analysis, and sometimes dialysis to ensure that conditions are right for the next column. Then, scientists must take their samples to the next column and begin the process anew.
Automating that purification process such that the eluate of interest from one column is shuttled automatically to the next column frees researchers from need frequent engagement with actual purification processes. An automated system can reduce small batch-to-batch variations between purification runs or those introduced when different individuals perform the work because the system reproducibly and repeatedly performs the same optimized method.
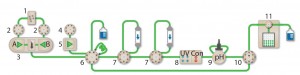
Figure 1: The NGC Discover 10 Pro system with an additional column switching valve illustrates the connectivity between modules used: components are 1 = buffer blending valve, 2 = buffer inlet valves, 3 = system pumps, 4 = sample inlet valve, 5 = sample pump, 6 = sample injection valve, 7 = column switching valves, 8 = multiwavelength detector with integrated conductivity monitor, 9 = pH module, 10 = outlet valve, 11 = BioFrac fraction collector.
Multidimensional (Multi-D) chromatography (Figure 1) on Bio-Rad’s Next Generation Chromatography (NGC) system automates all purification steps. Once a purification protocol is optimized, researchers push a button for continuous multistep purification. This method of automation is the only way to ensure consistent and reproducible purifications.
To illustrate the value of Multi-D chromatography, we focus here on two common applications for producing both native and recombinantly expressed proteins:
• screening aggregation levels of a monoclonal antibody (MAb) in tissue-culture fluid
• purification of a polyhistidine-tagged protein.
Automated Screening of Biologics for Aggregation
During biologics production, soluble aggregates can form, which poses a significant risk to patients. Bioprocessors must screen several conditions to eliminate or reduce those structures, typically using chromatography.
Aggregate screening begins with a researcher extracting and purifying a biologic from a tissue culture matrix using affinity chromatography, typically on a protein A column for MAbs. Size-exclusion chromatography (SEC) is used to resolve and quantify the ratio of monomer to aggregate.
A traditional workflow requires researchers to run samples on an affinity column, then neutralize and pool the fractions together before aggregation determination over an SEC column. In this case study, researchers simplified aggregate screening using automated Multi-D chromatography to combine column chemistries without the need to fractionate, neutralize, pool, and reload a sample between columns.
Methods: Bio-Rad scientists prepared human immunoglobulin G (IgG) MAb from Chinese hamster ovary (CHO) cells with three levels of aggregation: 0%, 10%, and 25%. They used a UNOsphere SUPrA affinity A (protein A) column (Bio-Rad Laboratories) for affinity purification, a Bio-Scale Mini Bio-Gel P-6 desalting column (Bio-Rad Laboratories) for buffer exchange (Multi-D workflow only), and an ENrich SEC 650 column (Bio-Rad Laboratories) for size exclusion.
Traditional Sequential Chromatography Workflow: Researchers loaded CHO cell lysate onto an affinity column, collected the elution fractions containing IgG, and neutralized them by adding Tris. They loaded pooled fractions onto an SEC column for aggregation analysis. This procedure was performed in triplicate on all three IgG samples containing the different aggregation levels.
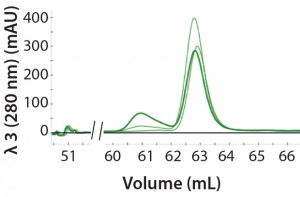
Figure 2: An overlay of three different samples on the SEC column containing three amounts of aggregation (0%, 10%, and 25%); peak height at 61 mL represents the degree of aggregation in the different samples.
Automated Multi-D Procedure: The automated procedure used an NGC system (Bio-Rad Laboratories) to elute IgG samples from an affinity column directly onto a desalting column for buffer exchange. Then the system routed the sample onto an SEC column for aggregate analysis (Figure 2).
Scientists loaded each IgG sample automatically through a sample inlet valve and integrated sample pump on the NGC system, which then automated their loading onto the columns. The system automatically cleaned the columns between samples.
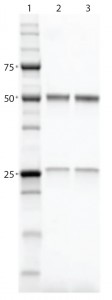
Figure 3: SDS-PAGE gel comparing purified proteins using both workflows; lane 1, Precision Plus Protein Unstained Standards; lane 2, traditional sequential chromatography eluate; lane 3, Multi-D chromatography eluate
Results and Discussion: Researchers performed purification and aggregate analysis in triplicate for each workflow and determined percent recovery and purity. The Multi-D workflow and the traditional sequential workflow demonstrated similar percent recovery. SDS-PAGE analysis of the IgG fractions collected after SEC showed that the two workflows purified the MAb equally well (Figure 3), although the automated Multi-D approach was achieved in a much shorter time (less than a day compared with two days for the traditional method). Figure 4 shows the Multi-D method’s high level of reproducibility, which overlays chromatograms of three repeat injections of the same sample.
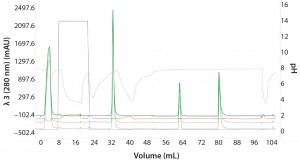
Figure 4: Three repeat injections of the same sample (monomer with 0% aggregation) using the sample pump to highlight the reproducibility of the automated Multi-D workflow
Automated Purification of a Polyhistidine-Tagged Protein Tagging proteins with a string of histidines is a popular molecular biology technique to facilitate protein purification using affinity chromatography. For maximum purity of a final product, many intermediate and polishing steps are conducted following initial Ni2+-immobilized metal affinity chromatography (IMAC). The added purity, however, comes with an added cost of time and labor. Purification of those proteins would benefit from a faster and more efficient process.
In a traditional sequential chromatography workflow, the eluate from an IMAC column is either diluted or dialyzed to lower its ionic strength before being loaded onto an ion-exchange (IEX) column for further purification. With the Multi-D method using the NGC system, researchers perform “dialyzation” using a desalting/buffer exchange column after the IMAC step and before the IEX step. So the final Multi-D method incorporates IMAC, desalting, IEX, and SEC in a single automated workflow.
Methods: To demonstrate efficacy of the Multi-D method, Bio-Rad’s researchers purified a common recombinant engineered protein called GFPsf, which is a superfolder green fluorescent protein with an ubiquitin-modifier at the N-terminus and polyhistidine at the C-terminus. Researchers expressed GFPsf in Escherichia coli and prepared a lysate to inject into the NGC system. Before creating a multicolumn purification method in NGC’s ChromLab software, each individual column step had to be optimized.
IMAC Capture Optimization: To understand the chromatography conditions best suited for purifying GFPsf, researchers used the NGC system’s “scouting” feature. This capability enables a sequence of purifications run under a range of conditions to determine the optimal setting for binding, washing, and elution. The system sequentially injects 10 samples of GFPsf lysate over an IMAC column (Bio-Scale Mini Nuvia IMAC column from Bio-Rad Laboratories), increasing the imidazole concentration of preelution wash steps by 5% increments from 0% to 45% buffer B (Tris HCl + 1M NaCl) each injection. Using this method, they identified the imidazole concentration for optimal contaminant removal and subsequently for elution of GFPsf. Data from this screening were used to generate the Multi-D method with the correct percentage of imidazole for the preelution wash buffer (10%) and elution volume.
Desalting and IEX Intermediate Optimization: After the IMAC column, researchers used the desalting template to run the GFPsf through a Bio-Scale Mini Bio-Gel P-6 desalting column to exchange the IMAC buffer with a low-ionic–strength buffer suitable for loading the protein onto the IEX column. Researchers then used the anionexchange (AEX) template to load the desalted GFPsf onto an ENrich Q anionexchange column.
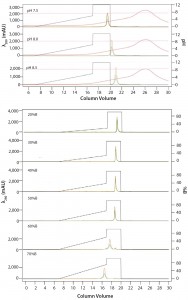
Figure 5: ENrich Q AEX chromatography scouting; a buffer blending module was used to generate the pH scouting runs using Tris buffer at pH 7.5, 8.0, and 8.5 for the ENrich Q column scouting. Tighter GFPsf binding is demonstrated by the increasing amounts of salt required to elute the protein as pH increases further from the theoretical protein pI of 6.2 (top). Using pH 8.0 from the pH scouting, the linear gradient %B endpoint was scouted against, starting at 20%B and increasing to 70%B in increments of 10%B. A GFPsf peak shift to the linear gradient begins at 50% and reaches completion by 70%B (bottom).
Researchers determined optimal binding and elution conditions for the AEX column again using the scouting feature within ChromLab software (Buffer A = Tris HCl) (Figure 5 top). They observed an increase in binding strength as the pH moved further away from the pI of the protein. The researchers scouted the final percentage of buffer B for the elution phase, screening a series of six runs from 20–70% B (Figure 5 bottom).
SEC Polish: For final polishing, researchers applied the GFPsf on a HiPrep Sephacryl S-300 HR SEC column (GE Healthcare) as the final column in the purification workflow.
Multi-D Method Development and Validation: For the Multi-D method, the researchers incorporated the optimal conditions from their method scouting to create a four-column automated Multi-D method. Figure 6 shows a run of the IMAC Multi-D purification workflow.
Results and Discussion: The final automated purification took about 12 hours to run, which is a significant time saving compared with the traditional sequential approach (which takes 3–4 days per sample). The Multi-D method also eliminates several labor-intensive steps typically required in between traditional chromatography steps. SDS-PAGE no longer needs to be run to visualize a protein after each step. The method also removes the need to dialyze IMAC eluate overnight to remove buffer salts and imidazole.
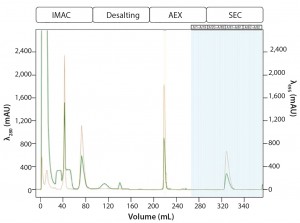
Figure 6: Automated Multi-D IMAC purification method; clarified GFPsf lysate (2 mL) was loaded onto a 5-mL Nuvia IMAC column using the sample pump. The column was washed with 10%B to remove contaminant proteins before step-elution from the column and storage in a 10-mL static loop. The target protein was loaded from the loop onto the 50-mL P-6 desalting column to remove the imidazole and lower the ionic strength of the buffer, then eluted isocratically and collected into a 20 ml static loop. The 20-mL protein sample was loaded onto a 1-mL ENrich Q column and eluted with a linear gradient. Using threshold collection, the eluate was shuttled into a 5-mL static loop, loaded onto a 120-mLl HiPrep Sephacryl S-300 HR column in a single injection, and eluted isocratically in 5-mL fractions.
Most of the time in a Multi-D run is spent on equilibration and elution from the SEC column. In this case, however, the time can be further reduced by performing multiple injections on a smaller, more pressure-tolerant SEC column such as the ENrich 70 or ENrich 650 columns (Bio-Rad Laboratories). Doing so could produce purified protein in about five hours. Another benefit of the automated method is increased reproducibility between purification batches by removing sources of potential human error. Ultimately, automated purification shows much promise for consistent protein production at laboratory scale.
Simplified Analysis
Bio-Rad’s NGC system allows for the easy adoption of Multi-D chromatography. The Multi-D workflow provides one-push functionality that enables users to start a method and walk away. The system is equipped with user-friendly software that simplifies the programming of customizable Multi-D workflows. Furthermore, the software enables easy and simultaneous analysis of multiple chromatography runs.
The automated nature of Multi-D chromatography also limits user error, enabling researchers to generate purified protein samples consistently and reproducibly.
Corresponding author Jeff Habel is collaborations and applications manager, Phillip Elms is senior scientist, Philip Chapman is global product manager, and Gunjan Choudhary is technical writer/editor at Bio-Rad Laboratories, 6000 James Watson Hercules, CA 94547; jeff_habel@bio-rad.com.
