The Chinese hamster ovary (CHO) cell line was first established by Theodore Puck in the 1950s and was used mainly for cytogenetics studies (1). Since the 1990s, CHO cells have increased in popularity as expression host cells because they can be adapted easily into suspension culture and genetically modified. The CHO cell line also has a human-like glycosylation profile (2–4).
Therapeutic proteins undergo different posttranslational modifications (PTMs) during manufacturing. Some modifications can lead to undesired effects such as decreased stability, loss of efficacy, and increased immunogenicity (5–7). Thus, controlling PTMs is a key task in process development to ensure that manufactured biologics have high and consistent quality.
Many chemical modifications such as oxidation, deamidation, glycation, and isomerization can be introduced during multiple manufacturing steps (8). However, other modifications are catalyzed by cellular enzymes expressed by production host cells. The most prominent example is glycosylation (Figure 1) (8). The structure or consensus motif of therapeutic proteins typically governs how they are modified enzymatically, but host gene expression and cell culture conditions also influence the extent of modifications significantly. Below, we focus on such enzyme-mediated PTMs identified in CHO culture and on corresponding control strategies.
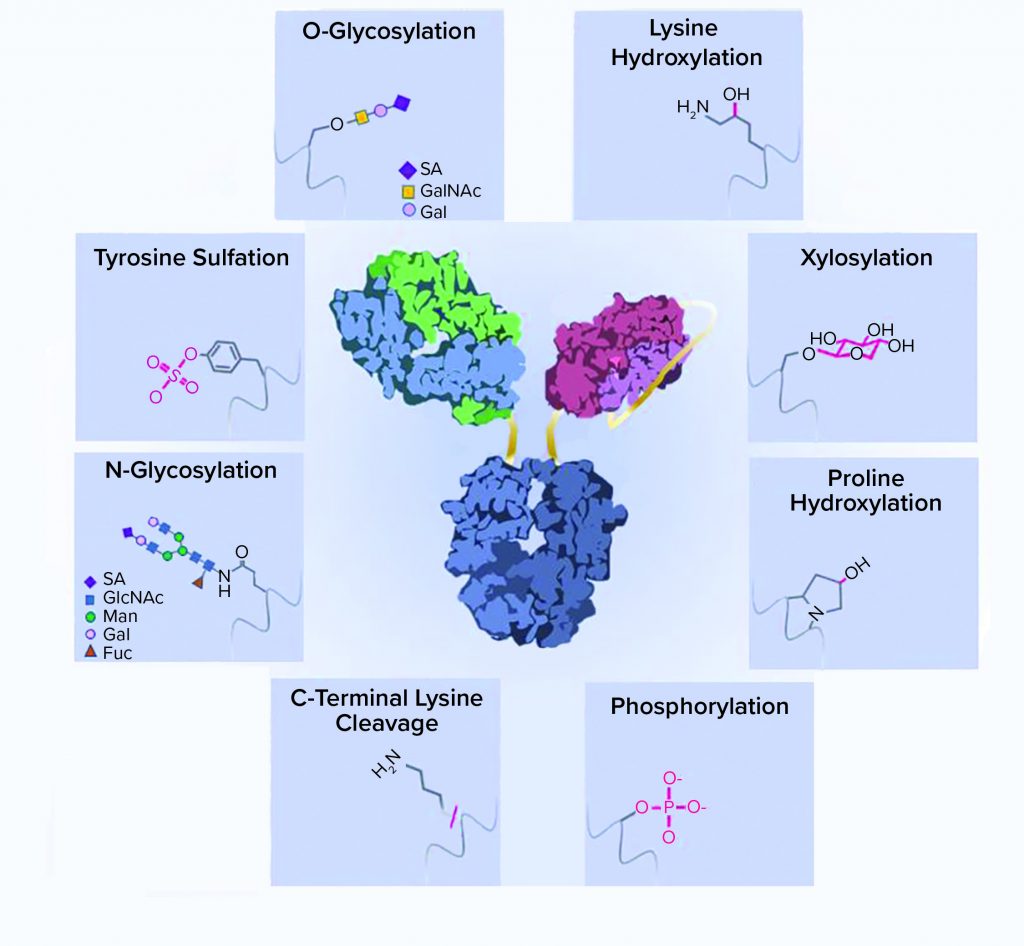
Figure 1: A schematic representation of recombinant-protein posttranslational modifications (PTMs); a typical bispecific antibody structure is shown as an example of a recombinant protein. Protein side chains are shown with corresponding modifications. ScFv = single chain Fv, SA = sialic acid, GalNAc = N-acetylgalactosamine, GlcNAc = N-acetylglucosamine, Man = mannose, Gal = galactose, Fuc = fucose
N-Glycosylation
Asparagine-linked glycosylation is a common PTM of recombinant proteins expressed by mammalian cells. The consensus sequence for N-glycosylation is Asn-Xaa-Ser/Thr (where Xaa is any amino acid except Pro), and the rarer sequence is Asn-Xaa-Cys (9, 10). N-linked oligosaccharides have a complex and versatile structure. They contain a common core with two N-acetylglucosamine (GlcNAc) and three mannose (Man) moieties. High-mannose glycans have more mannose moieties added to both branches of their cores. By contrast, complex glycans have GlcNAc added to both branches instead of mannose. Galactose (Gal) and terminal sialic acid (SA) can be further added to extend the oligosaccharide chain (Figure 1).
Hybrid glycans have one branch with mannose and another with GlcNAc added to their cores. A fucose (Fuc) can be added to the first GlcNAc in the core of complex and hybrid glycan structures. The relatively rare bisected glycans have an extra GlcNAc added onto the first mannose in the core and cannot be further fucosylated.
Among all rodent cell lines, CHO cells produce the most human-like glycans, with minimal levels of immunogenic epitopes such as galactose-α1,3-galactose (alpha-Gal) or N-glycolylneuraminic acid (Neu5Gc). Although glycan structures of CHO cells don’t have terminally linked α-2,6-sialic acids and bisecting GlcNAc, such glycans have relatively low abundance in human serum IgG. Thus, the effect of their absence on product quality and bioactivity is considered to be negligible (11, 12).
N-glycosylation is especially important for IgGs because the N-linked oligosaccharides present in the constant CH2 domain affect biological activity, protein conformation, stability, solubility, secretion, pharmacokinetics, and immunogenicity (13). Recombinant antibodies expressed in CHO culture contain predominantly fucosylated complex glycans with different levels of terminal galactose (G0F, G1F, and G2F) and some high-mannose structures. Glycosylated proteins containing high mannose glycan structures can be bound and cleared by mannose receptors (14), resulting in poor pharmacokinetics profiles. Similarly, glycosylated proteins with terminal galactose residues are cleared by asialoglycoprotein receptors (15), thereby rendering recombinant proteins with higher levels of sialylation longer plasma half-lives. However, only a small percentage of CHO-produced recombinant antibodies is sialylated, so the effect of sialylation on antibody clearance is minimal. Glycosylation of antibodies significantly influences their effector functions, including complement-dependent cytotoxicity (CDC) and antibody-dependent cellular cytotoxicity (ADCC). For example, high galactosylation leads to stronger C1q binding, thus higher CDC activity. A relatively lower degree of fucosylation improves the binding to CD16a expressed on natural killer cells and increases ADCC activity (16). Glycan structures also affect the thermostability of antibodies, with terminal galactosylated antibodies being the most stable form (16).
The control of N-glycosylation through CHO cell engineering has been studied extensively over the past few decades. For example, Yang et al. determined the function of 19 glycosyltransferases by knocking them out individually or in combination and further engineered CHO cell lines to produce proteins with homogenous N-glycans (17). The fucosylation of antibodies produced in CHO cells can be modulated by different cell engineering. Deletion of the Fut8 gene, which encodes only the fucosyltransferase responsible for fucosylation in CHO cells, has produced fully afucosylated antibodies with significantly increased ADCC activity (18, 19). Similar results were achieved by deleting Slc35c1, which encodes a GDP-fucose transporter (20).
Alternatively, antibodies without core fucose can be produced by CHO cells overexpressing the N-acetylglucos-aminyltransferase-III enzyme to catalyze the formation of bisected glycans, which cannot be fucosylated (21). Another creative way to generate afucosylated antibodies is to overexpress a prokaryotic enzyme GDP-6-deoxy-d-lyxo-4-hexulose reductase, which deflects the fucose de novo synthesis pathway, thus depleting endogenous fucose substrate (22).
Sialylation level also can be modulated by CHO cell engineering. For example, overexpression of enzymes involved in sialylation (including human β-galactoside α(2,6) sialyltransferase I, UDP-GlcNAc 2-epimerase/ManNAc kinase, and N-acetylglucosaminyltransferases I and IV) leads to significantly increased sialylation (23, 24).
Sialylation also can be improved by manipulating the expression of sialidases that catalyze the removal of sialic acids from a glycan structure. Sialidases are released into cell culture fluid upon cell membrane damage (25). The deletion of three sialidase genes (Neu1, 2, and 3) maintained the sialic acid content and the proportion of tetra-sialylated recombinant human erythropoietin (rhEPO) throughout fed-batch cultures without compromising cell growth and rhEPO production. Further knockout of apoptotic genes Bax and Bak improved productivity and increased dramatically the percentage of tetrasialylated recombinant protein. That increase probably was the result of less sialidase being released into the culture medium when cell viability improved (26).
N-glycosylation of recombinant proteins also can be modulated during production processes. For example, fucose analogs and their derivatives have high efficacy in inhibiting fucosylation (27, 28). The levels of galactosylation precursors galactose and uridine and galacosyltransferase cofactor manganese can be adjusted to modulate galactosylation (29, 30).
Sialylation can be improved by adding dexamethasone, which stimulates galactosyltransferase and sialytransferase expression (31). Sialylation also can be improved by controlling the feeding strategy of two nucleotide precursor sugars: glucosamine (GlcN) and N-acetylmannosamine (ManNAc) (32). In addition, 2,3-dehydro-2-deoxy-N-acetylneuraminic acid, a sialidase inhibitor, can reduce sialic acid removal, thereby increasing sialylation (33). Culture conditions such as temperature, pH, and osmolality also can influence the level of glycans such as high mannose (34).
O-Glycosylation
O-linked glycosylation is a common PTM found on recombinant proteins expressed by mammalian cells. The most common form of O-glycan is the mucin type, which contains an α-glycosidic linkage between N-acetylgalactosamine (GalNAc) (and sometimes GlcNAc) and the hydroxyl group of serine or threonine (35). No clear consensus sequence has been identified for O-glycosylation, so predicting O-glycosylation is difficult. Other moieties can be added onto GalNAc or GlcNAc to extend the O-glycan structures further. Those include GalNAc, galactose, GlcNAc, fucose, sialic acids, and the sulfo group (36). As a result, O-linked glycan structures could be more diverse than N-glycans.
Typically, O-linked glycosylation is observed on recombinant proteins, but it is rarely found on monoclonal antibodies. Valliere-Douglass et al. have reported on O-fucose modification with a net addition of 146Â Da in the complementarity determining region-1 (CDR-1) of one antibody light chain. In that study, the amino acid sequence around the modified Ser did not conform to known O-fucosylation sequence motifs (37).
Peptide linkers, which are used regularly to connect multiple protein domains, are key structural elements of multispecific antibodies and fusion proteins. Flexible linkers can minimize the steric interference between domains, but their high solvent accessibility increases susceptibility to cleavage and modification. Some studies have discovered the O-glycosylation (xylose modification) of linkers with glycine/serine (GS) repeats, which are the most commonly used peptide linkers (38, 39). Complex glycan structures also can form following initial placement of xylose on Ser (40), thus increasing the difficulty of controlling them during processing.
In some cases, O-glycosylation is important for the expression of recombinant proteins. For example, O-glycosylation can help the expression of human chorionic gonadotropin (hCG), a glycoprotein containing four O-glycosylation sites.Overexpressing uridine diphosphate-glucose pyrophosphorylase 2 (Ugp2, involved in O-glycan precursor synthesis) and polypeptide N-acetylgalactos-aminyltransferase 1 (Galnt1, involved in O-glycan sugar chain synthesis) increased O-glycosylation and total hCG protein expression significantly. Adding O-glycan precursors such as GalNAc to a culture medium improved hCG expression, presumably through increased O-glycosylation (41).
Tyrosine Sulfation and Other Modifications
Tyrosine sulfation is a PTM commonly found on secretory and plasma membrane proteins (42). Tyrosine sulfation facilitates protein–protein interactions. For example, sulfation of P-selectin glycoprotein ligand-1 (PSGL-1) enhances binding to the receptor P-selectin (43, 44). Two studies have focused on tyrosine sulfation on recombinant antibodies expressed in CHO cells (45, 46). In both cases, the modification occurred in the variable regions of antibody light chain. Because of potential potency and immunogenicity concerns, tyrosine sulfation must be controlled during drug substance manufacturing.
Our group found that differential expression of genes in the sulfation pathway led to significantly different tyrosine sulfation levels in different CHO hosts. Such genes include tyrosylprotein sulfotransferase 2 (TPST2) and the sulfation substrate transporter SLC35B2 (47). Thus, sulfation can be mitigated by host-cell selection using TPST2 and SLC35B2 as biomarkers or the modulation of the expression of both genes.
Our group also identified sodium chlorate as an effective sulfation inhibitor (>50% sulfation reduction). This inhibitor had no significant effect on CHO culture performance and other product quality attributes (47).
Other PTMs catalyzed by cellular enzymes in CHO cells include hydroxylysine, hydroxyproline, phosphorylation, and C-terminal lysine cleavage. Posttranslational hydroxylation of lysine typically is found on mammalian proteins such as collagen with a consensus sequence (XKG), catalyzed by lysyl hydroxylase complex. Xie et al. reported on the hydroxylation of a lysine on the heavy chain variable domain of a recombinant antibody (48). In that study, the consensus sequence alone was not sufficient for predicting whether the lysine would be modified because other lysines with XKG motif in the same antibody were not modified.
Peptide linkers are susceptible to more modifications than the xylosylation described above. For example, Spahr et al. found hydroxyproline on glycine-proline linker (49), and Tyshchuk et al. observed phosphorylation of serine on a short GS linker (50). Such relatively rare modifications pose challenges to process control, yet their biological and biophysical consequences still are unknown.
The cleavage of C-terminal lysine is another common modification on the heavy chain of recombinant antibodies produced in CHO cells. Hu et al. have reported that carboxypeptidase D is the only enzyme responsible for antibody C-terminal lysine cleavage in CHO cells (51). In that study, knockout of the gene in CHO cells completely abolished the C-terminal lysine cleavage of recombinant antibodies. However, C-terminal lysine is cleaved off rapidly in vivo, with a half-life of about an hour (52), thus the biological significance of this modification might be minimal.
Other PTMs such as oxidation, deamidation, and glycation can occur not only in cell culture, but also during other steps of a manufacturing process and drug product storage and handling (8, 53). The discussion of the control strategy of these modifications is beyond the scope of our review.
Mitigating Impacts of PTMs
PTM control during biopharmaceutical manufacturing often is challenging and resource-demanding. Thus, identifying problematic PTMs can be beneficial during the developability assessment stage and before process development (54). Selecting a final protein sequence with the most desired PTM profile can mitigate development risks significantly. Currently, the general practice is to produce protein materials of candidate sequences using a high-throughput transient expression and purification process followed by developability assessment using a panel of standard analytical methods, sometimes including stress conditions (55). That approach can assess a large number of sequences rapidly. However, the cell line and production process for developability assessment usually are vastly different from the ones that would be used for manufacturing.
It is well known that different host cells produce molecules with different levels of PTMs because of their intrinsic difference in genetic makeup. Some cryptic modifications with no consensus sequence can show up in one host but not another. Process conditions such as medium components and pH also can alter cellular enzymatic reactions and influence PTM profiles. Thus, product quality can differ greatly between materials used for developability assessment and those used in clinical studies. So using a scale-down manufacturing process to produce materials for candidate molecules is important. Once a problematic PTM (e.g., a cryptic glycosylation in the CDR) is identified, it can be eliminated through protein engineering. If the issue cannot be mitigated by protein engineering, a control strategy can be developed early in anticipation of potential development challenges.
Importance of PTM Control
PTMs of recombinant proteins can influence the biological activity, stability, pharmacokinetics, and immunogenicity of drug substances. Such modifications must be controlled during drug development to ensure the high quality and batch-to-batch consistency of clinical supplies. Some of the PTMs can be controlled in cell culture by CHO cell engineering and culture condition optimization. PTM control risk also can be mitigated by assessing the developability of candidate molecules at an early stage using materials produced with a scale-down manufacturing process.
References
1 Wurm MJ, Wurm FM. Naming CHO Cells for Biomanufacturing: Genome Plasticity and Variant Phenotypes of Cell Populations in Bioreactors Question the Relevance of Old Names. Biotechnol. J. 16(7) 2021: 2100165; https://doi.org/10.1002/biot.202100165
2 Hacker DL, De Jesus M, Wurm FM. 25 Years of Recombinant Proteins from Reactor-Grown Cells: Where Do We Go from Here? Biotechnol. Adv. 27(6) 2009: 1023–1027; https://doi.org/10.1016/j.biotechadv.2009.05.008
3 Zang M, et al. Production of Recombinant Proteins in Chinese Hamster Ovary Cells Using A Protein-Free Cell Culture Medium. Biotechnol. 13(4) 1995: 389–392; https://doi.org/10.1038/nbt0495-389.
4 Tihanyi B, Nyitray L. Recent Advances in CHO Cell Line Development for Recombinant Protein Production. Drug Discovery Today: Technologies 12 April 2021; https://doi.org/10.1016/j.ddtec.2021.02.003
5 Goetze AM, Schenauer MR, Flynn GC. Assessing Monoclonal Antibody Product Quality Attribute Criticality Through Clinical Studies. MAbs 2(5) 2010: 500–507; https://doi.org/10.4161/mabs.2.5.12897
6 Liu H, et al. Heterogeneity of Monoclonal Antibodies. J. Pharm. Sci. 97(7) 2008: 2426–2447; https://doi.org/10.1002/jps.21180.
7 Kuriakose A, Chirmule N, Nair P. Immunogenicity of Biotherapeutics: Causes and Association with Posttranslational Modifications. J. Immunol. Res. 2016: 1298473; https://doi.org/10.1155/2016/1298473
8 Liu H, et al. In Vitro and In Vivo Modifications of Recombinant and Human IgG Antibodies. MAbs 6(5) 2014: 1145–1154; https://doi.org/10.4161/mabs.29883.
9 Pless DD, Lennarz WJ. Enzymatic Conversion of Proteins to Gglycoproteins. Proc. Nat. Acad. Sci. 74(1) 1977: 134–138; https://doi.org/10.1073/pnas.74.1.134.
10 Satomi Y, Shimonishi Y, Takao T. N-Glycosylation at Asn491 in the Asn-Xaa-Cys Motif of Human Transferrin. FEBS Lett. 576(1–2) 2004: 51–56; https://doi.org/10.1016/j.febslet.2004.08.061.
11 Raju TS, et al. Species-Specific Variation in Glycosylation of IgG: Evidence for the Species-Specific Sialylation and Branch-Specific Galactosylation and Importance for Engineering Recombinant Glycoprotein Therapeutics. Glycobiol. 10(5) 2000: 477–486; https://doi.org/10.1093/glycob/10.5.477.
12 Hamako J, et al. Comparative Studies of Asparagine-Linked Sugar Chains of Immunoglobulin G from Eleven Mammalian Species. Comp. Biochem. Physiol. B 106(4) 1993: 949–954; https://doi.org/10.1016/0305-0491(93)90056-b.
13 Mattu TS, et al. The Glycosylation and Structure of Human Serum IgA1, Fab, and Fc Regions and the Role of N-Glycosylation on Fcα Receptor Interactions. J. Biol. Chem. 273(4) 1998: 2260–2272; https://doi.org/10.1074/jbc.273.4.2260
14 Goetze AM, et al. High-Mannose Glycans on the Fc Region of Therapeutic IgG Antibodies Increase Serum Clearance in Humans. Glycobiol. 21(7) 2011: 949–959; https://doi.org/10.1093/glycob/cwr027.
15 Ashwell G, Harford J. Carbohydrate-Specific Receptors of the Liver. Annu. Rev. Biochem. 51(1) 1982: 531–554; https://doi.org/10.1146/annurev.bi.51.070182.002531.
16 Wada R, Matsui M, Kawasak N. Influence of N-glycosylation on Effector Functions and Thermal Stability of Glycoengineered IgG1 Monoclonal Antibody with Homogeneous Glycoforms. MAbs 2019. 11(2) 2019: 350–372; https://doi.org/10.1080/19420862.2018.1551044.
17 Yang Z, et al. Engineered CHO Cells for Production of Diverse, Homogeneous Glycoproteins. Nat. Biotechnol. 33(8) 2015: 842–844; https://doi.org/10.1038/nbt.3280.
18 Malphettes L, et al. Highly Efficient Deletion of FUT8 in CHO Cell Lines Using Zinc-Finger Nucleases Yields Cells that Produce Completely Nonfucosylated Antibodies. Biotechnol. Bioeng. 106(5) 2010: 774–783; https://doi.org/10.1002/bit.22751.
19 Yamane-Ohnuki N, et al. Establishment of FUT8 Knockout Chinese Hamster Ovary Cells: An Ideal Host Cell Line for Producing Completely Defucosylated Antibodies with Enhanced Antibody-Dependent Cellular Cytotoxicity. Biotechnol. Bioeng. 87(5) 2004: 614–622; https://doi.org/10.1002/bit.20151.
20 Chan KF, et al. Inactivation of GDP-Fucose Transporter Gene (Slc35c1) in CHO Cells by ZFNs, TALENs and CRISPR-Cas9 for Production of Fucose-Free Antibodies. Biotechnol. J. 11(3) 2016: 399–414; https://doi.org/10.1002/biot.201500331.
21 Ferrara C, et al. Modulation of Therapeutic Antibody Effector Functions By Glycosylation Engineering: Influence of Golgi Enzyme Localization Domain and Coexpression of Heterologous β1, 4-N-Acetylglucosaminyltransferase III and Golgi α-Mannosidase II. Biotechnol. Bioeng. 93(5) 2006: 851–861; https://doi.org/10.1002/bit.20777.
22 von Horsten HH, et al. Production of Non-Fucosylated Antibodies By Coexpression of Heterologous GDP-6-Deoxy-d-lyxo-4-hexulose Reductase. Glycobiol. 20(12) 2010: 1607–1618; https://doi.org/10.1093/glycob/cwq109.
23 Thi Sam N, et al. Enhancement of Glycosylation by Stable Coexpression of Two Sialylation-Related Enzymes on Chinese Hamster Ovary Cells. J. Biosci. Bioeng. 126(1) 2018: 102–110; https://doi.org/10.1016/j.jbiosc.2018.01.010.
24 Cha H-M, et al. Cooverexpression of Mgat1 and Mgat4 in CHO Cells for Production of Highly Sialylated Albumin-Erythropoietin. Enzyme Microbial Technol. 103, 2017: 53–58; https://doi.org/10.1016/j.enzmictec.2017.04.010.
25 Chuan KH, et al. Caspase Activation, Sialidase Release and Changes in Sialylation Pattern of Recombinant Human Erythropoietin Produced by CHO Cells in Batch and Fed-Batch Cultures. Cytotechnol. 51(2) 2006: 67–79; https://doi.org/10.1007/s10616-006-9016-5.
26 Ha TK, et al. Knockout of Sialidase and Proapoptotic Genes in Chinese Hamster Ovary Cells Enables the Production of Recombinant Human Erythropoietin in Fed-Batch Cultures. Metabol. Eng. 57, 2020: 182–192; https://doi.org/10.1016/j.ymben.2019.11.008.
27 Okeley NM, et al. Development of Orally Active Inhibitors of Protein and Cellular Fucosylation. Proc. Nat. Acad. Sci. 110(14) 2013: 5404–5409; https://doi.org/10.1073/pnas.1222263110.
28 Allen JG, et al. Facile Modulation of Antibody Fucosylation with Small Molecule Fucostatin Inhibitors and Cocrystal Structure with GDP-Mannose 4,6-Dehydratase. ACS Chem. Biol. 11(10) 2016: 2734–2743; https://doi.org/10.1021/acschembio.6b00460.
29 Gramer MJ, et al. Modulation of Antibody Galactosylation Through Feeding of Uridine, Manganese Chloride, and Galactose. Biotechnol. Bioeng. 108(7) 2011: 1591–1602; https://doi.org/10.1002/bit.23075.
30 Prabhu A, Gadgil M. Nickel and Cobalt Affect Galactosylation of Recombinant IgG Expressed in CHO Cells. BioMetals 32(1) 2019: 11–19; https://doi.org/10.1007/s10534-018-0152-0.
31 Jing Y, Qian Y, Li ZJ. Sialylation Enhancement of CTLA4-Ig Fusion Protein in Chinese Hamster Ovary Cells By Dexamethasone. Biotechnol. Bioeng. 107(3) 2010: 488–496; https://doi.org/10.1002/bit.22827.
32 Cha H-M, et al. Nucleotide Sugar Precursor Feeding Strategy to Enhance Sialylation of Albumin-Erythropoietin in CHO Cell Cultures. Process Biochem. 66, 2018: 197–204; https://doi.org/10.1016/j.procbio.2017.12.014.
33 Gramer MJ, et al. Removal of Sialic Acid from a Glycoprotein in CHO Cell Culture Supernatant by Action of an Extracellular CHO Cell Sialidase. Biotechnol. 13(7) 1995: 692–698; https://doi.org/10.1038/nbt0795-692.
34 Mastrangeli R, et al. The Formidable Challenge of Controlling High Mannose-Type N-Glycans in Therapeutic MAbs. Trends Biotechnol. 38(10) 2020: 1154-1168; https://doi.org/10.1016/j.tibtech.2020.05.009.
35 Tejwani V, et al. Glycoengineering in CHO Cells: Advances in Systems Biology. Biotechnol. J. 13(3) 2018: 1700234; https://doi.org/10.1002/biot.201700234.
36 Olson, FJ, et al. A MUC1 Tandem Repeat Reporter Protein Produced in CHO-K1 Cells Has Sialylated Core 1 O-Glycans and Becomes More Densely Glycosylated If Coexpressed with Polypeptide-GalNAc-T4 Transferase. Glycobiol. 15(2) 2004: 177–191; https://doi.org/10.1093/glycob/cwh158.
37 Valliere-Douglass JF, et al. O-Fucosylation of an Antibody Light Chain: Characterization of a Modification Occurring on an IgG1 Molecule. Glycobiol. 19(2) 2008: 144–152; https://doi.org/10.1093/glycob/cwn116.
38 Wen D, et al. Discovery and Investigation of O-Xylosylation in Engineered Proteins Containing a (GGGGS)n Linker. Anal. Chem. 85(9) 2013: 4805–4812; https://doi.org/10.1021/ac400596g.
39 Spahr C, et al. Recombinant Human Lecithin-Cholesterol Acyltransferase Fc Fusion: Analysis of N- and O-Linked Glycans and Identification and Elimination of a Xylose-Based O-Linked Tetrasaccharide Core in the Linker Region. Protein Sci. 22(12) 2013: 1739–1753; https://doi.org/10.1002/pro.2373.
40 Spahr C, Shi SDH, Lu HS. O-Glycosylation of Glycine-Serine Linkers in Recombinant Fc-Fusion Proteins. MAbs 6(4) 2014: 904–914; https://doi.org/10.4161/mabs.28763.
41 Deng Z, et al. A Study on Enhanced O-Glycosylation Strategy for Improved Production of Recombinant Human Chorionic Gonadotropin in Chinese Hamster Ovary Cells. J. Biotechnol. 306, 2019: 159–168; https://doi.org/10.1016/j.jbiotec.2019.10.006.
42 Moore KL. The Biology and Enzymology of Protein Tyrosine O-Sulfation. J. Biol. Chem. 278(27) 2003: 24243–24246; https://doi.org/10.1074/jbc.R300008200.
43 Kehoe JW, Bertozzi CR. Tyrosine Sulfation: A Modulator of Extracellular Protein-Protein Interactions. Chem. Biol. 7(3) 2000: R57–R61; https://doi.org/10.1016/s1074-5521(00)00093-4.
44 Rodgers SD, Camphausen RT, Hammer DA. Tyrosine Sulfation Enhances But Is Not Required for PSGL-1 Rolling Adhesion on P-Selectin. Biophys. J. 81(4) 2001: 2001–2009; https://doi.org/10.1016/S0006-3495(01)75850-X.
45 Zhao J, et al. Characterization of a Novel Modification of a CHO-Produced MAb: Evidence for the Presence of Tyrosine Sulfation. MAbs 9(6) 2017: 985–995; https://doi.org/10.1080/19420862.2017.1332552.
46 Tyshchuk O, et al. Characterization and Prediction of Positional 4-Hydroxyproline and Sulfotyrosine, Two Posttranslational Modifications That Can Occur at Substantial Levels in CHO Cells-Expressed Biotherapeutics. MAbs 11(7) 2019: 1219–1232; https://doi.org/10.1080/19420862.2019.1635865.
47 LiuR, et al. Modulating Tyrosine Sulfation of Recombinant Antibodies in CHO Cell Culture By Host Selection and Sodium Chlorate Supplementation. Biotechnol. J. 16(9) 2021: 2100142; https://doi.org/10.1002/biot.202100142.
48 Xie Q, Moore B, Beardsley RL. Discovery and Characterization of Hydroxylysine in Recombinant Monoclonal Antibodies. MAbs 8(2) 2016: 371–378; https://doi.org/10.1080/19420862.2015.1122148.
49 Spahr C, et al. High-Resolution Mass Spectrometry Confirms the Presence of a Hydroxyproline (Hyp) Posttranslational Modification in the GGGGP Linker of an Fc-Fusion Protein. MAbs 9(5) 2017: 812–819; https://doi.org/10.1080/19420862.2017.1325556.
50 Tyshchuk O, et al. Detection of a Phosphorylated Glycine-Serine Linker in an IgG-Based Fusion Protein. MAbs 9(1) 2017: 94–103; https://doi.org/10.1080/19420862.2016.1236165.
51 Hu Z, et al. Carboxypeptidase D Is the Only Enzyme Responsible for Antibody C-Terminal Lysine Cleavage in Chinese Hamster Ovary (CHO) Cells. Biotechnol. Bioeng. 113(10) 2016: 2100–2106; https://doi.org/10.1002/bit.25977.
52 Cai B, Pan H, Flynn GC. C-Terminal Lysine Processing of Human Immunoglobulin G2 Heavy Chain In Vivo. Biotechnol. Bioeng. 108(2) 2011: 404–412; https://doi.org/10.1002/bit.22933.
53 Xu P, et al. Applications of Small Molecules in Modulating Productivity and Product Quality of Recombinant Proteins Produced Using Cell Cultures. Biotechnol. Adv. 43, 2020: 107577; https://doi.org/10.1016/j.biotechadv.2020.107577.
54 Xu Y, et al. Structure, Heterogeneity and Developability Assessment of Therapeutic Antibodies. MAbs 11(2) 2019: 239–264; https://doi.org/10.1080/19420862.2018.1553476.
55 Bailly M, et al. Predicting Antibody Developability Profiles Through Early Stage Discovery Screening. MAbs 12(1) 2020: 1743053; https://doi.org/10.1080/19420862.2020.1743053.
Yixiao Zhang is senior scientist, Zhimei Du is executive director, and corresponding author Ren Liu is principal scientist at Process Cell Sciences, Merck & Co., Inc., 2000 Galloping Hill Road, Kenilworth, NJ 07933.; ren.liu@merck.com.

