Pandemics such as the current COVID-19 outbreak pose tremendous healthcare and economic challenges. Vaccines hold promise for controlling pandemics; however, substantial challenges come with pandemic-response vaccine development, manufacturing, distribution, and administration. To address those now, many companies are using rapid-response vaccine-production platform technologies. Computational modeling tools could help further accelerate development of those technologies, increase production and distribution efficiencies, and reduce costs and risks once vaccine platforms are fully developed and validated. To those ends, a set of modeling methodologies — including bioprocess modeling for quality by design (QbD), technoeconomic, and supply chain modeling planning — can be combined with prior knowledge and current data from underlying physical processes. The results provide a framework to accompany and enhance vaccine production and distribution processes.
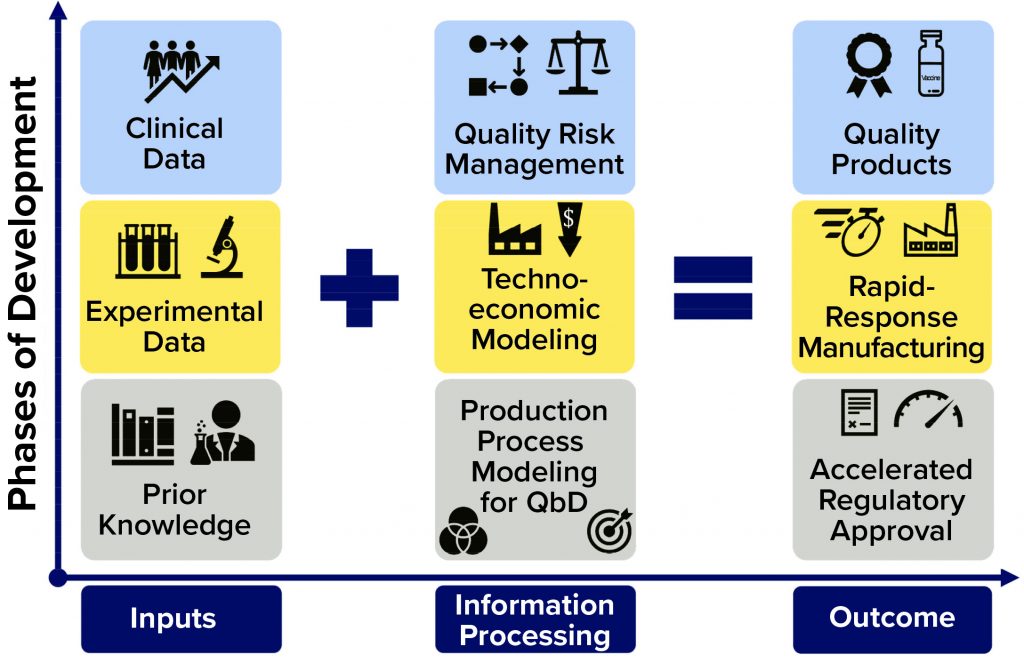
Figure 1: Framework for further accelerating regulatory approval and pandemic-response manufacturing of high-quality vaccines; on the horizontal axis, the flow of information is shown (from inputs through information processing to outputs); on the vertical axis, the overall phases of development are indicated. The information processing phases (middle column) can be carried out in parallel, with iterative communication between bioprocess modeling for quality by design (QbD), technoeconomic modeling, and quality risk assessment. Platform technologies that use the same production process to make a wide range of vaccines against different diseases can benefit the most from this framework.
Pandemic-Response Vaccine Challenges
The scale and urgency of vaccine development during a pandemic caused by a new disease present unprecedented difficulties. They put enormous strain on the entire industry, from preclinical testing and clinical trials to procurement of raw materials and consumables, drug-substance and drug-product manufacturing, quality control, distribution, and administration to the human population (1–5). To overcome some of those challenges, clinical testing and regulatory processes are fast-tracked, and phases of clinical trials are carried out in parallel (1–5). Thus, COVID-19 vaccine development is happening at record speed, about 10-fold faster than the pace of conventional vaccine development (1).
Manufacturing at pandemic scales and rates also poses an extraordinary challenge because there are no dedicated facilities for COVID-19 or “Disease X” vaccine production (6). Existing facilities must be adapted and new processes established for pandemic vaccine production (1, 2, 4). Rapid-response manufacturing platform technologies are being used for achieving that, but never have been used at this scale (1, 3). Some modalities, such as RNA vaccine platforms, had not produced a licensed vaccine product before December 2020 (1, 3). Thus, production-platform technologies are being developed and optimized even while they are used to produce vaccine-candidate materials for clinical trials.
During a pandemic, supply chains can be disrupted by transportation restrictions and sourcing limitations for raw materials, consumables, and access to specialized facilities (e.g., for animal testing) (7, 8). Once vaccines are produced — depending on their thermal stability — product distribution also could be challenging (1). Some vaccines (e.g., RNA vaccines) need to be stored at –70 °C during each step in transportation and storage. Thus, in the worst-case scenario, distribution to low- and middle-income tropical and subtropical countries becomes extremely challenging because those lack appropriate cold-chain infrastructures (1). Reduced thermostability also would limit the use of multidose vials and consequently increase drug-product manufacturing costs for single-dose format (or low dose numbers) as well as distribution costs because of cold-chain requirements (1). To solve this problem, some vaccine developers are assessing formulations for thermostability and/or considering lyophilized formulations (1).
Administration to large human populations could pose a bottleneck based on the time required to log each person, administer a vaccine dose, and observe patients after immunization to address rare cases of anaphylaxis. Throughout that process, virus spread among patients and between medical personnel and patients also must be prevented. Meanwhile, antivaccine movements could hinder developed countries’ abilities to achieve herd immunity (9, 10). “Vaccine-nationalistic” politics could prevent vaccine delivery to developing countries where they could be needed most, ultimately allowing disease to spread, increase mortalities, and prolong disruptions in the global economy (11, 12).
However, even if all those challenges can be resolved — and if through all remarkable efforts and technological developments unprecedented development timelines are achieved — vaccines cannot be supplied fast enough to stop the spread of the current pandemic and reduce related mortalities in progress. Thus, the industry needs to accelerate vaccine development further while reducing risks associated with clinical development, manufacturing, supply chains, and vaccination campaigns.
Rapid-Response Platforms
Rapid development, manufacturing, distribution, and administration of high-volume vaccines are crucial for overcoming pandemics. Rapid-response technologies such as RNA, DNA, and adenovirus vector platforms are being used for SARS-CoV-2 vaccine production (13). Such technologies can produce a wide range of vaccine candidates against different diseases using the same production, purification, and formulation process with the same quality control approach. Thus, once they have been developed and validated fully, these platforms can produce vaccines against new and currently unknown diseases — e.g., “Disease X” (6) — some 10× faster than conventional vaccine production technologies (1). Drug-substance manufacturing costs could be well below US$1/dose, depending mostly on the RNA amount per dose (to be determined during clinical trials), production scales (influenced by economies of scale), and the type and size of vials/containers (which depend on the thermostability of liquid or lyophilized products) (1).
Specifically, two broad types of RNA vaccines are in development based on messenger (mRNA) and self-amplifying (saRNA) platforms. In both cases, vaccine antigens are produced inside a recipient’s own cells, based on genetic instructions delivered by the mRNA or saRNA vaccine sequence. However, saRNA vaccines can replicate themselves in human cells, so they are estimated to require ≤100-fold smaller doses than mRNA vaccines (14, 15). That can make 100× faster drug-substance production possible in terms of doses per batch and thus lower the cost per dose for saRNA compared with mRNA vaccines.
At the Future Vaccine Manufacturing Hub led by Imperial College London, we are developing four outbreak-response manufacturing platform technologies: saRNA products; virus-like particles (VLPs) with genetically customizable surface epitopes produced using the baculovirus expression vector system (BEVS) with insect cells; humanized, glycoengineered yeast for high-yield expression of recombinant proteins; and bacterial outer-membrane vesicles with genetically customizable membrane antigens (16). All of these technologies facilitate production of a wide range of vaccine candidates against different diseases using the same basic production processes (16).
Three of the platforms have produced candidates against SARS-CoV-2, with the saRNA vaccine candidate entering clinical trials in June 2020. In early December 2020, the Pfizer/BioNTech mRNA vaccine gained regulatory approval in the United Kingdom, and the Moderna followed soon after. In addition, four replication-deficient adenovirus-vector candidate vaccines (in the United Kingdom, the Russian Federation, the United States, and China), four inactivated viral-vaccine candidates (three in China and one in India), and one protein subunit candidate vaccine (in the United States) were in late-phase clinical trials (13). In early phase trials are other RNA and DNA platform-produced vaccines as well as recombinant-protein and inactivated-virus candidates (13).
Transformative, rapid-response RNA platform technologies can produce vaccine candidates quickly because (1)
- they are “agnostic” to infectious-disease targets
- they use simple, enzymatic production processes with just a few unit operations
- they have short batch-cycle times of one to two days, depending on scheduling
- they offer high productivity per unit volume and time.
For example, the saRNA platform can produce several million doses’ worth of drug substance each day per liter of bioreactor working volume, depending mostly on the RNA amount required for each vaccine dose — currently being determined in clinical studies (1). Once these platforms have been developed and validated fully, vaccines against new and existing diseases will be able to move quickly through the development and manufacturing pipelines (1).
Further Accelerating Vaccine Production
To accelerate vaccine deployment even more, production platforms can be complemented by computational modeling and quality risk assessment tools based on QbD (17, 18) and technoeconomic assessment principles (1, 19, 20). The models proposed below would incorporate prior knowledge, results from preclinical and clinical trials, and production-process data. Input information can be processed by the modeling framework to ensure product quality, increase production volumes and rates, reduce costs, and (in principle) speed up the clinical testing and regulatory approval process (Figure 1).
Figure 1’s vertical axis shows the phases of development, and the horizontal axis illustrates the flow of information. Because the output properties of the RNA polymer do not change significantly from one disease target to the next, the same modeling framework can be combined with that vaccine platform irrespective of the target. Thus, information obtained from development and production of any RNA vaccine can be used to improve the modeling framework performance and consequently enhance the production processes. Once fully developed and validated, and in synergy with the platform technologies (e.g., the RNA vaccine platform), this framework would accelerate production of a wide range of vaccine candidates against most disease targets (including “Disease X”), ultimately improving process performance and enabling rapid-response, high-volume production of a wide range of vaccines at high quality and low cost. Such modeling frameworks also could improve the robustness, stability, and reproducibility of manufacturing while reducing material, labor, and energy consumption. For QbD, a design space can be defined to keep critical process parameters (CPPs) within regulatory limits and achieve target product critical quality attributes (CQAs).
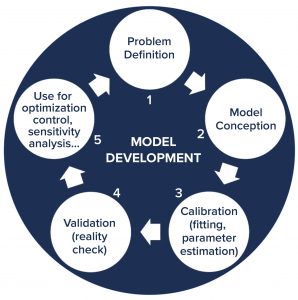
Figure 2: General approach for developing computational models; the development cycle starts with understanding and defining the problem to be solved. Next, the model is conceptualized and then calibrated with experimental data. Following that, the model is validated and then implemented to serve its purpose (such as to optimize process conditions, control the production process, or perform uncertainty and sensitivity analysis). Throughout its use, the model can be improved to provide more advanced process understanding, which leads to better understanding of the system and the underlying problem. Then that could initiate a new iteration in the model development cycle.
Model Development
QbD bioprocess models can be statistical and data-driven (e.g., based on linear regression, principal component analysis, partial least squares regression, machine learning, or artificial neural networks) or mechanistic (e.g., based on enzymatic or growth kinetics), or they can combine both in hybrid models (1). Each model is constructed starting from an understanding of the problem to be solved (Figure 2). Following its conception, a model is calibrated with experimental data — also called model fitting mostly for statistical models, parameter estimation for mechanistic models, and model training in machine-learning models. Next, a model is validated with additional experimental data to determine whether it can predict what is happening in real-world systems. Prior knowledge and data from unsuccessful experiments (not producing desired outcomes) are needed for model calibration and validation.
Following validation, a model is implemented, usually either to support process development or operation — e.g., by relating CQAs of the product to CPPs of its manufacturing process (1). Models can be used to optimize conditions, control processes (e.g., by model-predictive control), and carry out sensitivity analyses to determine what uncertainties or variations in input parameters most affect product quality attributes and key performance indicators. Thus, sensitivity analysis also can be used to identify CPPs.
As models are used, they can provide increased process understanding and in turn be improved based on the data they are fed with and the scenarios they assess. Thus, the development cycle enters a new iteration, starting again from problem understanding. In data-poor environments, mechanistic models tend to have the highest predictive power by incorporating prior knowledge in the form of mechanistic process understanding. As large amounts of data become available in later development stages, data-driven models can outperform mechanistic models, and in many cases, hybrid models are preferable.
Modeling Approaches
Models can be used throughout process development, manufacturing, and distribution stages in varying degrees of connectivity with physical processes, as Figure 3 shows. The solid arrows indicate strong connections between models, making it possible either to repurpose or adapt them or to enable large amounts of information to flow between them. The dashed arrows show weak connections between models, with limited information flow. Applying a comprehensive modeling framework to all stages of development can increase vaccine deployment speeds substantially while ensuring high quality and reducing cost.
That way, as depicted on the x-axis of Figure 3, early on in process development and scale-up, a factorial design of experiments (DoE) (21) or model-based DoE (MBDoE) approach can be used to maximize the information gleaned from experimental data (22, 23). This would require carrying out experiments that emphasize connectivity with the physical (laboratory or pilot scale) process, shown in the top blue row of Figure 3. Resulting data can be used for calibrating and validating bioprocess models for QbD with intermediate coupling to the physical process, shown in the middle yellow row of Figure 3. Because data generated through DoE are used for QbD modeling, the links between these two are strong, as illustrated by the solid black arrows.
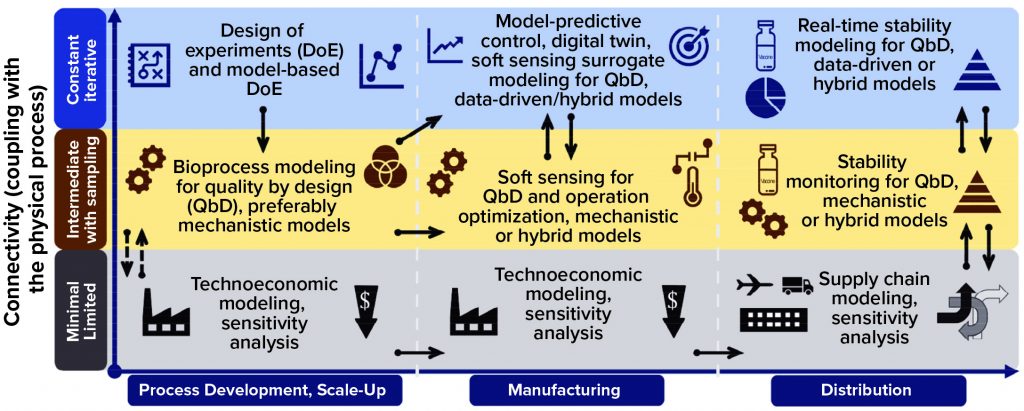
Figure 3: Computational modeling approaches for aiding vaccine deployment; models are shown for the following three stages of deployment across the horizontal axis (process development and scale-up, manufacturing, and distribution). For each stage, models are tiered by their level of connectivity with the underlying production process or distribution network. Thus, minimal connectivity is represented in the bottom (gray) row, intermediate connectivity is represented in the middle yellow row, and high connectivity is represented in the top blue row. The black arrows indicate interaction and information flow between the different modeling approaches, with solid arrows showing strong and dashed arrows showing weak interaction or information flow.
Developing mechanistic models can be advantageous at the start of new-product development, when limited data are available and if the process is understood mechanistically (e.g., based on enzymatic kinetics, cell growth, or protein expression). For platform technologies, with which the same process produces a wide range of products, prior knowledge from previous products also can be used for developing the next product to overcome data scarcity.
Early on, whole-process flow models (technoeconomic models) can be built based on process understanding and knowledge of similar processes, with minimal direct connectivity to a laboratory or pilot-scale process — as Figure 3 shows in gray (bottom row). In general, such models require minimal interfacing with bioprocess models for QbD, as indicated by the dashed black arrows. Using whole-process models, stochastic-uncertainty and global-sensitivity analyses can be carried out to assess how uncertainty propagates through a model and evaluate to what degree key performance indicators are affected by uncertainty ranges in model inputs.
Manufacturing: At high, constant, iterative connectivity with the physical manufacturing process (center column, top blue row), a model-predictive controller (digital twin) would take in real-time measurement data from the process and predict near-future values (e.g., 5–10 minutes ahead) for key performance indicators and CQAs based on those data (24–26). If deviations outside predefined acceptable product-CQA ranges are predicted, the model would the model would take corrective measures (control actions) and adjust CPPs to correct for those deviations and faults in product CQAs before they occur — with minimal or no human involvement (24–26).
Such a model-predictive controller can help to automate a process, reduce human error, and operate the process within a set design space for QbD, thus keeping CPPs within limits to obtain target CQAs. Process fluctuations caused by inherent variability in input materials and biological systems can be solved in real-time to maintain consistent product CQAs in desired ranges. A model-predictive controller could be generated in principle by simplifying the QbD bioprocess model from process development and scale-up, as indicated by the solid black arrows. That would provide a surrogate model that could run constantly in parallel with the manufacturing process (real time).
Those models also can be used for soft sensing, in which a model takes in easy-to-obtain measurement data from a physical (“hard”) sensor and computes data (e.g., concentrations or CQA values) that would be difficult or expensive to measure in real time. With data obtained through the model (software), these are called soft sensors (27, 28). For example, the concentration of a reaction product or cell metabolite can be calculated using a model of reaction or cellular metabolism kinetics and experimental measurements of a reactant or cell-nutrient input concentration (27, 28). This indirect determination of a reaction-component concentration can be useful when it cannot be measured directly and/or such measurements would be expensive and/or time-consuming.
If a soft sensor takes in real-time measurements constantly using, for example, on-line and in-line process analytical technology (PAT), the model needs also to provide solutions in real time. Surrogate, data-driven, statistical, or hybrid models can be used; mechanistic models that usually solve differential equations are slower and are not suitable for this application. If data are obtained by sampling (e.g., using at-line and off-line PAT) or calculated from other datasets, then direct connectivity to the process decreases (Figure 3, yellow row). In such a case, the models need not be fast or run in real time, so mechanistic models can be used. Data and model features can be transferred between highly and intermediately process-connected models, as indicated by the solid black arrows in Figure 3.
Overall, soft sensors can be extremely valuable for solving challenges in the bioprocessing industry related to the lack of adequate PAT. A soft sensor also can become part of a vaccine production platform together with the QbD modeling framework. With low process connectivity — in gray, bottom row of the figure — the technoeconomic models can be similar to their counterparts from the earlier development phase, as indicated by the solid black arrows. Technoeconomic models can be used during manufacturing, for example, for scheduling and operational optimization as well as simulating production campaigns and scenarios for meeting predefined demand and cost targets.
| Useful Software Packages |
| DoE can be carried out using JMP from the SAS Institute, R from the R Foundation, Matlab from MathWorks, or packages (e.g., pyDOE) for Python programming, among other options. MBDoE can be carried out using Python packages and libraries, gProms from Process Systems Enterprise Limited, or Matlab. Mechanistic bioprocess modeling and stability modeling for QbD, surrogate modeling for model-predictive control, and soft-sensing modeling can be performed using Python packages and libraries or in the Matlab program. Data-driven (e.g., machine learning and artificial neural-network modeling) and hybrid modeling usually are performed using Python packages and libraries. Technoeconomic modeling can be performed in SuperPro Designer from Intelligen or in BioSolve Process from Biopharm Services, for example. Supply chains can be modeled in the open-source Hermes program or (if mathematical optimization and/or additional customization are required) in GAMS from the GAMS Development Corporation. Global sensitivity and uncertainty analysis can be carried out using Solbol GSA available from Imperial College London or using Sobol, Halton, Latin hypercube, or random sampling toolboxes in Matlab or other software packages. |
In the distribution stage (right column of Figure 3), real-time stability models for QbD are in constant, iterative connection with the physical process (a series of storage and transportation steps). These models would compute the real-time degradation of vaccines as a function of real-time temperature and time measurements (29–32). First, a model describing the appropriate physical, chemical, and/or biological degradation kinetics (33, 34) needs to be conceptualized, calibrated, and validated. In chemical degradation cases, for example, the kinetic rate constant can be linked with temperature by the Arrhenius equation, describing the proportion of degraded vaccines as a function of temperature and time for a wide range of temperature–time profiles in addition to a limited set of prevalidated conditions (29–32). Because these models would run in real time, they require fast solutions, such as statistical, data-driven, surrogate, and hybrid models; computationally intensive mechanistic models would not work for this application.
Stability modeling for QbD also can be carried out with intermediate connectivity to the underlying process, which consists of a series of storage and transportation steps. In this case, as shown in the yellow row of Figure 3, the QbD stability modeling could be used to compare different scenarios and optimize delivery options in combination with supply-chain models. Therefore, the solid black arrows indicate a close connection among all the levels of connectivity in the distribution deployment stage. With the least direct connectivity to the actual underlying storage and transportation processes, supply-chain models also take in information from the technoeconomic models in the manufacturing stage, as illustrated by the solid black arrows. The distribution network and routes can be optimized mathematically to increase delivery rates and to reduce costs and risks. Moreover, if QbD stability and supply-chain models are linked, then a supply chain can be optimized to maximize stability and remaining shelf life of a vaccine.
By applying a holistic modeling framework with models connected to underlying production and distribution processes and interconnected with each other, overall vaccine supply rates and volumes can be increased, product quality can be maintained at a high level, and vaccination costs can be reduced. This approach could prove to be extremely valuable for future outbreaks once these vaccine production platform technologies and modeling frameworks are fully developed, validated, and integrated.
Facilitating Further Acceleration
The world urgently needs large amounts of vaccines at consistently high-quality to address pandemics. Technologies such as RNA platforms promise to produce large amounts of vaccine substances rapidly and at low cost. However, current fast-tracked timelines for clinical testing and regulatory approval are not short enough to stop the currently unfolding COVID-19 pandemic. To accelerate development of high-quality vaccines further, a modeling framework could accompany rapid-response vaccine production technologies. Such a framework integrates prior knowledge with data from preclinical and clinical testing and from manufacturing processes. Models would be built in a stepwise way to create an overall framework for the entire pipeline, from process development and scale-up through manufacturing to distribution. Such a framework could increase vaccine supply rates and volumes, help ensure the quality of vaccines produced, and reduce their costs.
Acknowledgments
This research is funded by the Department of Health and Social Care using UK Aid funding and managed by the Engineering and Physical Sciences Research Council (EPSRC, grant number EP/R013764/1). Views expressed herein are those of the author and not necessarily the Department of Health and Social Care. Funding is thankfully acknowledged from UK Research and Innovation (UKRI) through EPSRC grant number EP/V01479X/1 on COVID-19/SARS-CoV-2 vaccine manufacturing and supply chain optimization. The author gratefully acknowledges Professor Nilay Shah and Dr. Cleo Kontoravdi for helpful discussions and support of this work.
References
1 Kis Z, et al. Rapid Development and Deployment of High-Volume Vaccines for Pandemic Response. J. Adv. Manuf. Proc. 2(3) 2020: e10060; https://doi.org/10.1002/amp2.10060.
2 Furlong A. Europe’s Challenge of a Lifetime: Manufacturing Enough Coronavirus Vaccines. Politico SRL 2020; https://www.politico.eu/article/europes-challenge-of-a-lifetime-manufacturing-enough-coronavirus-vaccines.
3 Mullard A. COVID-19 Vaccine Development Pipeline Gears Up. Lancet 395(10239) 2020: 1751–1752; https://pubmed.ncbi.nlm.nih.gov/32505245.
4 Lurie N, et al. Developing COVID-19 Vaccines at Pandemic Speed. N. Engl. J. Med. 382(21) 2020: 1969–1973; https://doi.org/10.1056/NEJMp2005630.
5 Le TT, et al. The COVID-19 Vaccine Development Landscape. Nat. Rev. Drug Discov. 19, May 2020: 305–306; http://dx.doi.org/10.1038/d41573-020-00073-5.
6 Simpson S, et al. Disease X: Accelerating the Development of Medical Countermeasures for the Next Pandemic. Lancet Infect. Dis. 20(5) 2020: e108–15; https://doi.org/10.1016/S1473-3099(20)30123-7.
7 West S, et al. Is the World Ready to Produce a Billion Doses of a COVID-19 Vaccine? Imp. Coll. London – Eng. News 2020: https://www.imperial.ac.uk/news/197321/is-world-ready-produce-billion-doses.
8 Bozzay E, Cahen M. Stocktaking Report on Immediate Public Procurement and Infrastructure Responses to COVID-19. Organisation for Economic Cooperation and Development: Paris, France, 2020; https://www.oecd.org/coronavirus/policy-responses/stocktaking-report-on-immediate-public-procurement-and-infrastructure-responses-to-covid-19-248d0646.
9 Ball P. Anti-Vaccine Movement Could Undermine Efforts to End Coronavirus Pandemic, Researchers Warn. Nature 2020: 251; https://www.nature.com/articles/d41586-020-01423-4.
10 Johnson NF, et al. The Online Competition Between Pro- and Anti-Vaccination Views. Nature 582(7811) 2020: 230–233; https://doi.org/10.1038/s41586-020-2281-1.
11 Weintraub R, Bitton A, Rosenberg ML. The Danger of Vaccine Nationalism (Internet). Harv. Bus. Rev. 2020; https://hbr.org/2020/05/the-danger-of-vaccine-nationalism.
12 Milne R, Crow D. Why Vaccine ‘Nationalism’ Could Slow Coronavirus Fight. Financial Times 2020; https://www.ft.com/content/6d542894-6483-446c-87b0-96c65e89bb2c.
13 Draft Landscape of COVID-19 Candidate Vaccines: 12 November 2020. World Health Organization: Geneva, Switzerland, 2020; https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines.
14 McKay PF, et al. Self-Amplifying RNA SARS-CoV-2 Lipid Nanoparticle Vaccine Induces Equivalent Preclinical Antibody Titers and Viral Neutralization to Recovered COVID-19 Patients. bioRxiv 1 January 2020: 2020.04.22.055608; http://biorxiv.org/content/early/2020/04/25/2020.04.22.055608.abstract.
15 Moderna Advances Late-Stage Development of Its Vaccine (mRNA-1273) Against COVID-19. Moderna, Inc., 2020; https://investors.modernatx.com/news-releases/news-release-details/moderna-advances-late-stage-development-its-vaccine-mrna-1273.
16 Kis Z, et al. Emerging Technologies for Low-Cost, Rapid Vaccine Manufacture. Biotechnol. J. 14(1) 2019: 1800376; https://doi.org/10.1002/biot.201800376.
17 CMC-Vaccines Working Group. A-Vax: Applying Quality By Design to Vaccines. Parenteral Drug Association: Bethesda, MD, 2012; https://www.dcvmn.org/IMG/pdf/a-vax-applying-qbd-to-vaccines_2012.pdf.
18 Haas J, et al. Implementation of QbD for the Development of a Vaccine Candidate. Vaccine 32(24) 2014: 2927–2930; http://www.sciencedirect.com/science/article/pii/S0264410X14002011.
19 Kis Z, et al. A Model-Based Quantification of the Impact of New Manufacturing Technologies on Developing Country Vaccine Supply Chain Performance: A Kenyan Case Study. J. Adv. Manuf. Proc. 1(3) 2019: e10025; https://doi.org/10.1002/amp2.10025.
20 Petrides D. SuperPro Designer User Guide: A Comprehensive Simulation Tool for the Design, Retrofit, and Evaluation of Specialty Chemical, Biochemical, Pharmaceutical, Consumer Product, Food, Agricultural, Mineral Processing, Packaging and Water Purification. Wastewater: Scotch Plains, NJ, 2013; http://www.intelligen.com/downloads/SuperPro_ManualForPrinting_v10.pdf.
21 Antony J. Design of Experiments for Engineers and Scientists, Second Edition. Elsevier Science: Waltham, MA, 2014; https://books.google.co.uk/books?id=p7pCAgAAQBAJ.
22 Galvanin F, Barolo M, Bezzo F. A Framework for Model-Based Design of Experiments in the Presence of Continuous Measurement Systems. IFAC Proc. Vol. 43(5) 2010: 571–576; http://www.sciencedirect.com/science/article/pii/S1474667016300970.
23 Galvanin F, et al. A General Model-Based Design of Experiments Approach to Achieve Practical Identifiability of Pharmacokinetic and Pharmacodynamic Models. J. Pharmacokinet. Pharmacodyn. 40(4) 2013: 451–67; https://doi.org/10.1007/s10928-013-9321-5.
24 Kroll P, et al. Model-Based Methods in the Biopharmaceutical Process Lifecycle. Pharm. Res. 34(12) 2017: 2596–2613; https://pubmed.ncbi.nlm.nih.gov/29168076.
25 Sommeregger W, et al. Quality By Control: Towards Model Predictive Control of Mammalian Cell Culture Bioprocesses. Biotechnol. J. 12(7) 2017: 1600546; https://doi.org/10.1002/biot.201600546.
26 Mesbah A, et al. Model Predictive Control of an Integrated Continuous Pharmaceutical Manufacturing Pilot Plant. Org. Process Res. Dev. 21(6) 2017: 844–854; https://doi.org/10.1021/acs.oprd.7b00058.
27 Randek J, Mandenius C-F. On-Line Soft Sensing in Upstream Bioprocessing. Crit. Rev. Biotechnol. 38(1) 2018: 106–121; https://doi.org/10.1080/07388551.2017.1312271.
28 Luttmann R, et al. Soft Sensors in Bioprocessing: A Status Report and Recommendations. Biotechnol. J. 7(8) 2012: 1040–1048; https://doi.org/10.1002/biot.201100506.
29 Clénet D, et al. Advanced Kinetic Analysis As a Tool for Formulation Development and Prediction of Vaccine Stability. J. Pharm. Sci. 103(10) 2014: 3055–3064; http://www.sciencedirect.com/science/article/pii/S0022354915303749.
30 Clénet D. Accurate Prediction of Vaccine Stability Under Real Storage Conditions and During Temperature Excursions. Eur. J. Pharm. Biopharm. 125, 2018: 76–84; http://www.sciencedirect.com/science/article/pii/S0939641117308731.
31 Higashikawa F, Chang L-J. Kinetic Analyses of Stability of Simple and Complex Retroviral Vectors. Virology 280(1) 2001: 124–131; http://www.sciencedirect.com/science/article/pii/S0042682200907438.
32 Waterman KC. The Application of the Accelerated Stability Assessment Program (ASAP) to Quality by Design (QbD) for Drug Product Stability. AAPS PharmSciTech 12(3) 2011: 932; https://doi.org/10.1208/s12249-011-9657-3.
33 Ausar S, et al. Forced Degradation Studies: An Essential Tool for the Formulation Development of Vaccines. Vaccine Dev. Ther. 3, June 2013:11; http://www.dovepress.com/forced-degradation-studies-an-essential-tool-for-the-formulation-devel-peer-reviewed-article-VDT.
34 Manning MC, et al. Stability of Protein Pharmaceuticals: An Update. Pharm. Res. 27(4) 2010: 544–575; https://doi.org/10.1007/s11095-009-0045-6.
Zoltán Kis, PhD, is a research associate in the Centre for Process Systems Engineering, part of the Department of Chemical Engineering at Imperial College London, South Kensington Campus, London SW7 2AZ; z.kis10@imperial.ac.uk; http://www.imperial.ac.uk/people/z.kis10.
