Biopharmaceutical companies are racing to develop vaccines that mitigate the COVID-19 pandemic, taking a wide range of vaccine-development approaches that include traditional modalities and cutting-edge technologies based on DNA and RNA. Vaccine developers are leveraging robust manufacturing concepts and integrated processes to shorten timelines. Advanced analytics also are playing a critical role in ensuring the safety and efficacy of those emerging vaccines.
A New Wave of Vaccines
Vaccines based on attenuated viruses entail development timelines ranging from four years to 15 years (1). Thus, despite that approach’s proven efficacy, it is inappropriate for rapid responses. A virus must be isolated from hosts, a reliable culture system must be established, and the pathogen must be weakened chemically or biologically to ensure patient safety while eliciting sufficient immune responses. Finally, adjuvants must be optimized for final dosing. All those steps require validation using several analytical techniques and bioassays.
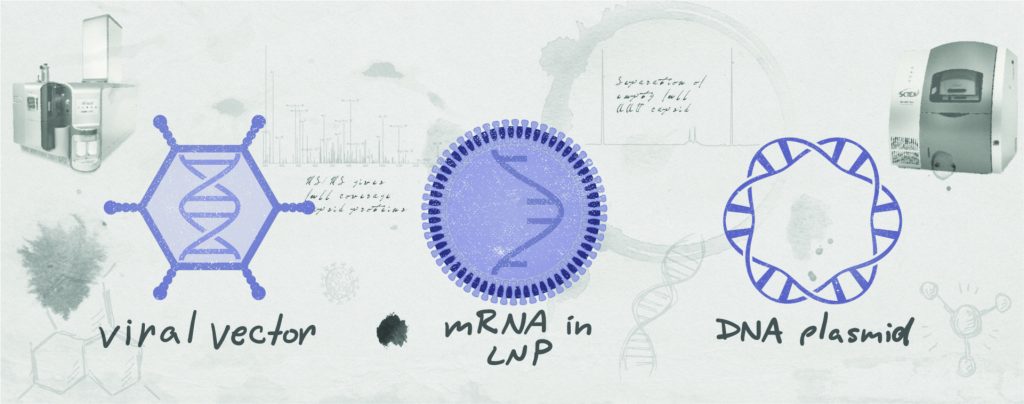
Emerging genetic vaccines eliminate the complexity and risk of working with live viruses. Approaches including those based on viral vectors, DNA plasmids, and messenger RNA (mRNA) produce viral antigens inside patient cells. Leveraging native protein translation and posttranslational-modification machinery within the host’s actual cells eliminates concerns about achieving efficient antigen uptake. And unlike attenuated and subunit vaccines that require a new process for each vaccine, emerging technologies require only the genetic sequence for parts of a target virus. Because formulation, production, packaging, and even safety profiles are nearly identical across different vaccines, robust, scalable platform processes can be developed to reduce timelines.
Plasmid-DNA Platforms: The first “naked-DNA” vaccines were developed and studied in the early 1990s. Since then, five plasmid-DNA products have received regulatory approval. Of those, four have veterinary indications; the fifth is a cancer vaccine for human use. Many plasmid-DNA vaccines, cancer therapies, gene therapies, and immunomodulatory treatments for autoimmune and allergic diseases are under development for use in humans. For instance, clinical trials of DNA vaccines have demonstrated boostable immune responses to Ebola and Marburg viruses, and the first two Zika vaccines to reach clinical development were DNA vaccines.
Inovio is a leader in developing DNA-based drug products, including vaccines for human papilloma virus (HPV), Ebola, Middle East respiratory syndrome (MERS), Zika, Lassa fever, and novel coronavirus disease 19 (COVID-19) as well as cancer vaccines and immunotherapies. The company has reached phase 2 of its planned phase 2/3 clinical trial of INO-4800 candidate, a room-temperature–stable plasmid-DNA vaccine candidate designed to protect against the sudden acute respiratory syndrome novel coronavirus (SARS-CoV-2).
Viral-vector vaccines use engineered viruses to enter host cells and deliver DNA that yields expression of tailored antigenic proteins. Many such vaccines have been approved for veterinary applications, and the European Medicines Agency (EMA) approved the first viral-vector vaccine regimen for humans in May 2020: the Ad26.ZEBOV and MVA-BN-Filo combination developed against Ebola virus by Janssen/Johnson & Johnson (J&J). Several other candidates have reached clinical studies for human immunodeficiency virus, respiratory syncytial virus (RSV), Zika, malaria, tuberculosis, MERS, rabies, cancer, COVID-19, and several rare diseases. Viral vectors also are used to deliver gene and gene-modified cell therapies, including several approved products and hundreds in clinical development.
Several vaccines against SARS-CoV-2 are based on viral vectors, including candidates from CanSino Biologics (adenovirus type 5, Ad5-nCoV), J&J (adenovirus subtype 26, JNJ-78436735), AstraZeneca (AZ)–Oxford University (chimpanzee-derived adenoviral vector, AZD1222), and ReiThera (previously Okairos, gorilla-derived adenoviral vector, GRAd-COV2). CanSino is conducting phase 3 studies in Russia, Pakistan, and Mexico. J&J expects to release its COVID-19 vaccine in the second half of 2021. AZ announced in November 2020 that AZD1222 is ~70% effective with two full doses but could be ~90% effective with a half- or full-dose regimen.
Vaccines based on mRNA use lipid nanoparticles (LNPs) to deliver mRNA templates into host cells, resulting in expression of viral antigens. Although naked RNA was shown to stimulate in vivo expression of encoded proteins during the early 1990s, immunogenicity has been a problem. Today, the stability and translational capacity of mRNA have improved, and immunogenicity concerns have been reduced by modifying specific nucleosides.
Several companies are developing mRNA-based drug products, including prophylactic vaccines for Zika virus, human metapneumovirus and parainfluenza type 3 (hMPV/PIV3), pediatric RSV, multiple influenza strains, Epstein-Barr virus, rabies, Lassa fever, yellow fever, malaria, and COVID-19. Companies also are leveraging mRNA for cancer vaccines and immunotherapies as well as therapies for cystic fibrosis, primary ciliary dyskinesia (PCD), pulmonary arterial hypertension (PAH), and idiopathic pulmonary fibrosis (IPF).
Multiple mRNA vaccines against SARS-CoV-2 have entered or moved beyond clinical trials. Pfizer and partner BioNTech received emergency use authorization (EUA) from the US Food and Drug Administration (FDA) for its BNT162 vaccine, with phase 3 results indicating ~90% effectiveness. Moderna has reported similar effectiveness for its mRNA-1273 product, which also has received EUA. Meanwhile, CureVac’s CVnCoV candidate began phase 2a testing in Panama and Peru in September 2020. Translate Bio anticipates entering the clinic with its own product early in 2021.
Advanced Analytics Beget Vaccine Development Success
Vaccine technologies have developed at an amazing pace during the pandemic. However, none of these platforms would have succeeded without comprehensive analytical support.
For both naked-DNA and viral-vector vaccines, plasmid purity and structure must be confirmed because those factors influence product efficacy, safety, and quality. Plasmids must be free of contaminants and have the right form (>80% supercoiled for bulk release), length, and sequence.
For viral vectors, expression of capsids with the correct size, peptide sequence, and posttranslational modifications is essential. Analysts must ensure capsid purity with respect to host-cell proteins (HCPs) and other genetic contaminants to minimize immunogenicity and off-target effects. Reduction of empty and partial capsids is equally important because low numbers of fully packaged vectors result in low infectivity and thus low antigen production.
Vaccines based on mRNA require similar analyses. In addition to verifying the correct length of mRNA, scientists must confirm the presence of a 5′ cap and a sufficiently long polyadenosine (polyA) tail to ensure in vivo stability and function. Because mRNA vaccines are formulated as fusogenic LNPs comprising cationic lipids, assessment of LNP formation is required to ensure effective mRNA delivery.
Effective Analytical Solutions
Many analytical technologies developed originally for protein therapeutics have been modified to address assessment requirements raised by emerging genetic vaccines.
Assessing Plasmid Purity: Capillary electrophoresis with laser-induced fluorescence detection (CE-LIF) provides rapid, sensitive, reproducible, and automated quantitative analysis of plasmid-DNA isoforms, ensuring plasmid purity. Supercoiled, linearized, and open-circular isoforms for 5-kb and 7-kb plasmids can be separated reproducibly with baseline resolution in under 15 minutes. Automated Sanger genome sequencing can simplify analysis of DNA in plasmid vectors.
The Sciex GenomeLab GeXP genetic-analysis system supports two unique chemistries — deoxyinosine triphosphate (dITP) and deoxyguanosine triphosphate (dGTP) — for routine and difficult analyses, respectfully. The latter assays include assessments of repeat regions, polymerase hardstops or regions rich in guanine–cytosine base pairings.
Evaluating Viral Vectors: New approaches are accelerating viral-vector analysis while still ensuring assay accuracy. Quadrupole time-of-flight mass spectrometry (Q-ToF MS) can characterize adenoassociated virus (AAV) capsid proteins rapidly. Proprietary Sciex SWATH (sequential window acquisition of all theoretical mass spectra) technology for peptide mapping can be combined with BioPharmaView Flex software to gather and analyze data for low-abundance peptides and PTMs (e.g., glycopeptides, deamidation sites, and disulfide bonds) that information-dependent peptide-mapping workflows can miss. All those data can be gleaned from a single MS sample injection.
SWATH-based liquid chromatography (LC)-MS/MS can identify and quantify thousands of HCPs and other contaminants in a single run. Meanwhile, CE–sodium dodecyl sulfate (SDS) improves upon traditional SDS-polyacrylamide gel electrophoresis (PAGE) for purity analysis of AAV capsids. Sample preparation and protein separation can be automated to reduce labor. Compared with SDS-PAGE, CE-SDS exhibits higher sensitivity for the low concentrations of viral proteins found in AAV samples as well as better resolution and quantitation capability. Good linearity of absorbance response to sample concentration means that CE-SDS also enables better assay reproducibility.
Denaturing agarose gel electrophoresis and Southern blot methods typically are used to assess accurately the quality and length/size of the genome encapsidated within a viral vector, but such assays are time consuming and offer limited resolution in size determination. Sciex has shown that CE-LIF enables rapid, repeatable, and accurate determination of AAV capsid size and purity. That method also can detect intact and partial AAV genomes as well as minute impurities.
Assays for mRNA: mRNA vaccines require similar analyses to those used for DNA and viral-vector vaccines. SWATH-based LC-MS/MS can confirm antigen sequences and identify off-target protein expression, whereas rapid and reproducible detection, separation, and sizing of mRNA can be achieved using CE-LIF with single-base resolution down to 15 nucleotides.
Because the 5′ cap and 3′ polyA-tail length are essential to mRNA stability and function, analysts must evaluate those components. After ribonuclease H (RNaseH) digestion, SWATH-based LC-MS/MS and CE-LIF should provide high-resolution analysis of 5′ RNA fragments of 5–40 bases and 3′ RNA fragments with polyA fragments of ~0–200 bases. Those methods are known to be effective, respectively, for analyzing synthetic antisense oligonucleotides of 15–20 bases and single-base resolution of the 40–60 polyA base standard.
Confirmation of mRNA-vaccine LNP composition can be achieved using a LC-MS/MS workflow supported by Sciex’s targeted lipidomic assay. Combined with the Scheduled MRM (multiple reaction monitoring) Pro algorithm and fast polarity switching, that assay enables identification and quantification of ~1,150 polar and neutral lipids.
A Committed Analytical Partner
Regardless of vaccine technology, it is essential to confirm that a desired antigen is expressed and that adequate T-cell responses are achieved. Sciex’s state-of-the-art MS, CE-LIF, and multiplex genome analyses are available to support those efforts. Visit https://info.sciex.com/vaccines to discover ways to accelerate vaccine development by getting the right answers the first time using precision analytics.
Reference
1 Cagle T. Until Now, What’s the Quickest a Vaccine Has Ever Been Developed? Nautilus 28 May 2020; https://coronavirus.nautil.us/until-now-whats-the-quickest-a-vaccine-has-ever-been-developed.
Corresponding author Susan Darling is senior director of product and market management for CE biopharmaceutical applications (susan.darling@sciex.com), and Todd Stawicki is global marketing manager for LC-MS biopharmaceutical applications at AB Sciex LLC, 1201 Radio Road, Redwood City, CA 94065; https://www.sciex.com.