Mammalian and microbial protein production platforms have been used for over 30 years to produce a number of successful biologic drugs, including monoclonal antibodies (MAbs), recombinant proteins, and therapeutic enzymes (1). Most biologics are produced by mammalian cell lines, with Chinese hamster ovary (CHO) cells being the most widely used. However, microbial cells also are used to express recombinant therapeutic proteins, and almost 30% of currently approved biologics are produced by Escherichia coli bacteria (2). With worldwide biologics sales >56 billion Euros per year (3), both industrial and academic bioprocess scientists have to develop methods for effective process development of such proteins to make them competitively priced and maintain profitability in this lucrative market.
One strategy is to work with user-friendly single-use, miniaturized bioreactors to shorten process development times. That increases efficiency and reduces the sources of human errors. The current challenge is to ensure that such minibioreactors can be used as scale-down models for larger scale microbial cultivations while improving laboratory efficiency and productivity.
Process Development
To define optimal media or process conditions, process development typically is performed using benchtop bioreactors after successful strain selection. Benchtop bioreactors can model pilot- and production-scale cultivations because they mimic the sparging and stirring principle of those larger scale reactors. Because benchtop bioreactors generally are made of glass, they require long set-up times and labor-intensive pre- and postprocess cleaning and sterilization with every experimental run. Additionally, benchtop models range from 1 L to 10 L in scale, so they required fairly large amounts of media and feeds to be used during each experiment. These bioreactors and the sensors they contain generally are reusable, so they always carry a risk of contamination and inconsistencies in sensor performance from run to run, leading to data inconsistencies and requiring several repeat runs to produce trustworthy results.
Limited time and budgets often restrict the use of benchtop bioreactors, preventing researchers and process engineers from running full design of experiments (DoE) testing to determine the optimum process conditions using these types of bioreactors. Sometimes that can lead to a suboptimal final choice of process conditions on scale-up, which ultimately will adversely affect yield and productivity. If more DoE runs could be performed under conditions that represent scale-up environments, then it would be possible to determine truly optimum cultivation conditions for use in biomanufacturing, derisking the development process and potentially saving thousands of Euros in manufacturing cost of goods (CoGs).
Automated Single-Use Bioreactors
For a single-use minibioreactor to be effective as a model for process development and process transfer, it must provide comparable mixing, gas sparging, and sampling parameters. Especially for microbial fermentation processes (for example, with E. coli), developing an appropriate single-use minibioreactor has been more challenging than for eukaryotic cells because bacteria cells reach high cell densities very rapidly. Hence, such a bioreactor must provide high oxygen transfer capacity, excellent mixing performance, and sufficient heat dissipation.
To meet those diverse requirements, Sartorius-Stedim Biotech developed the advanced miniature-scale ambr 250 bioreactor system in 2013. Since then, it has been demonstrated to be an efficient high-throughput process model for Pichia pastoris, E. coli, and CHO cells (4, 5). To broaden access to this technology, the ambr 250 modular system — a more compact, expandable version of the same technology with many features of the high-throughput system — was introduced in 2016.

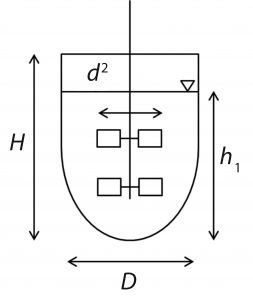
Photo 2: An ambr 250 modular vessel for microbial fermentation features feed reservoirs (chilled) including feeding ports, baffles, two six-blade Rushton impellers, a single-use pH sensor, and an integrated fluorescent DO sensor.
As shown in Photo 1, the new technology is an innovative new high-performance benchtop bioreactor system for parallel microbial or cell culture in 100-mL to 250-mL single-use vessels. This represents a unique, single-use, scale-down model for rapid process development and optimization. The basic design includes two bioreactors, and it can be expanded to configurations for four, six, or eight miniature bioreactors. All bioreactor stations have complete individual process control of all parameters including pH, dissolved oxygen (DO), temperature, and agitation.
Each single-use bioreactor is integrated fully with sensors, five reagent reservoirs, and syringe pumps to facilitate experimental set-up and turnaround. Units come equipped with an off-gas measurement system (Photo 2). In addition, the bioreactor vessels are geometrically similar to standard benchtop bioreactors, enabling the ambr 250 modular system to serve as a scale-down model. All this means that it helps bioprocess engineers increase their productivity significantly while eliminating the need for sterilizing the bioreactors or their accessories.
 Case Study: Performance of an E. Coli Cultivation
Case Study: Performance of an E. Coli Cultivation
The ambr 250 modular system comes in two configurations — one for eukaryotic cell culture, and one for microbial fermentation — with relevant stirrer configurations, gassing ranges, and so on. The aim of our study was to compare the performance of the ambr 250 modular system with that of an established UniVessel glass 5-L bioreactor for a high–cell-density E. coli cultivation. Our approach was first to address process engineering characterization and second to run a cultivation and compare the performance of both systems.
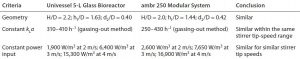
Table 1: Process engineering characterization of an ambr 250 modular system and a UniVessel glass 5-L reactor
Process Engineering Characterization: For process engineering characterization of both systems (Table 1), we determined the geometries, kLa values, and power inputs using methods published by DECHEMA (6). Results indicated a large similarity, so with the same cultivation conditions, we could expect similar cultivation results.
Cultivation: To benchmark the ambr 250 modular system, we conducted a high–cell-density fed-batch cultivation of E. coli W3110 (DSM: 5911) and compared the results to data derived from a UniVessel glass 5-L cultivation. The “Cultivation Parameters” box lists conditions for both.
| Cultivation Parameters |
| The batch medium, fed-batch medium, and exponential feeding profile is as published by Korz DJ, et al. Simple Fed-Batch Technique for High Cell Density Cultivation of Escherichia coli. J Biotechnol. 39(1) 1995: 59–65). |
| Temperature: 37 °C. |
| pH Setpoint: controlled at 6.8 with a 20% ammonia solution |
| pO2 Setpoint: 20% with air and pure oxygen |
| Maximal Gassing Rate: 1 vvm |
| Vessel Geometry: 2 Ă— 6-blade disc impeller and baffles |
| Stirrer and Tip Speed: (ambr 250 modular system) ≤4,400 rpm, 4.6 m/s, (UniVessel glass 5-L bioreactor) ≤1,500 rpm, 5.0 m/s |
| Specific Growth Rate: μ setpoint of 0.15 h–1 |
To save time in process development, the ambr 250 modular system has to model microbial cultivation conditions of a benchtop bioreactor accurately. We chose an E. coli cultivation for this case study because microbial cultivations are more difficult to mimic than mammalian bioprocesses, especially with their more challenging requirements related to oxygen transfer and temperature control.
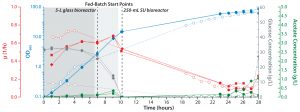
Figure 1: Overview of the E. coli cultivation in an ambr 250 modular system (two runs, diamonds and squares) in comparison with a process performed in a UniVessel glass 5-L reactor (circles); shown is the optical density at 600 nm (blue), growth rate (red), acetate (green), and glucose concentration (gray). Start of the feed is indicated by a dashed line.
To determine how closely the minibioreactor could model a benchtop bioreactor, we cultured E. coli W3110 (DSM: 5911) cells in chemically defined medium developed by Biener, et al. (7) with a fed-batch process for 28 hours at 37 °C. Stirrer tip speeds were 4.6–5.0 m/s in an ambr 250 modular system and a 5-L benchtop bioreactor, respectively. We analyzed cell growth, acetate and glucose concentrations, and we profiled pO2, temperature, and pH (Figures 1 and 2). Feed 1 started after the batch phase and feed 2 after cell densities reached 150 OD600.
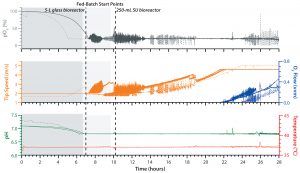
Figure 2: Comparing process control of both E. coli cultivations in an ambr 250 modular system (solid lines) in with a process performed in a UniVessel glass 5-L reactor (broken lines); shown is the pO2 (gray), which was controlled by changing the tip speed (orange) and supplementing oxygen (blue). Also controlled were pH (green) and temperature (red).
After 28 hours, the ambr 250 modular fed-batch cultivation achieved a final optical density of 335 OD600, which is equal to a final dry cell weight (DCW) of 120Â g/L. That is comparable to cell densities achieved by the UniVessel Glass 5-L reactor (Figure 1), which indicates that high cell density was reached within the ambr 250 modular system. Also, results from both types of vessel showed that pO2 was maintained at its setpoint of 20% throughout cultivation (Figure 2).
We measured a maximal oxygen uptake rate (OUR) of 600 mmol/L/h. By contrast with the kLa value determined during process engineering characterization (Table 1), we calculated the process kLa value >870 h–1 based on the pO2 and off-gas data (not shown). The reason for that increased kLa is the high response time of our pO2 sensor in the buffer system used during process engineering characterization compared with data derived from a cultivation.
Extracellular concentrations of glucose revealed that it had been completely metabolized during the batch phase, so the cells remained carbon limited during the fed-batch phase. Acetate was formed with ≤0.4 g/L glucose during the batch phase in both cultivations. Before fed-batch started, glucose was consumed completely and remained <1.0 g/L until the culture’s end in both types of bioreactor. The acetate formed remained below the inhibitory concentration of 5.0 g/L (8).
All other cultivation conditions were comparable for the two systems and remained in desired ranges. Consequently, no limitations occurred even at high cell densities, particularly regarding oxygen transfer and heat generation (Figure 2). Taking the geometric similarity and comparable power input under consideration, the ambr 250 modular system exhibited the same performance as that of an established glass bioreactor, so high–cell-density E. coli cultivations could be realized using the minibioreactors.
Case Study: Productivity
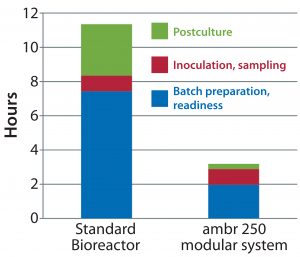
Figure 3: Timeline for pre- and postrun preparation of a 5-L glass benchtop bioreactor and the ambr 250 modular system
After we established that performance of the ambr 250 modular system is similar to that of a 5-L benchtop bioreactor, scientists at a leading contract research organization (CRO) compared the preparation time taken before and after culture runs in the benchtop 5-L glass bioreactor and the ambr 250 modular system. Their results (Figure 3) show that with reusable benchtop bioreactors, preparation and set-up took about 11 hours, whereas with the ambr 250 modular system each run took just three hours in total.
Based on those results, a scientist could prepare up to four single-use bioreactors per day on average — compared with setting up and cleaning a single glass benchtop bioreactor every two days. Replacing reusable benchtop bioreactors with single-use minibioreactors would increase process development capacity by up to fourfold.
Comparability Demonstrated
Single-use minibioreactor technology can provide comparable control of pO2, pH, and temperature in E. coli cultivations to that achieved with benchtop bioreactors. Even high–cell-density E. coli cultivations with high growth rates can be conducted and controlled without limitations. Setting up and running reusable benchtop bioreactors is manually intensive; the fully automated single-use minibioreactors are more convenient by comparison, taking much less time to set up and run and eliminating the need for postrun cleaning and sterilization. Therefore, process development and process transfer can be performed rapidly and efficiently with the minibioreactor system, increasing the number of cultivation runs that can be evaluated.
Those benefits improve the chances for process engineers to determine optimal bioprocessing parameters for maximum growth and protein expression in microbial production systems. The automated ambr 250 modular system enables DoE implementation into their work flows. Finally, the geometric similarity of these platforms reduces risk in scaling up to larger single-use pilot- and manufacturing-scale stirred bioreactors, which reduces timelines and provides significant cost savings related to materials and labor.
References
1 Johnson IS. Human Insulin from Recombinant DNA Technology. Science 219(4585) 1983: 632–637.
2 Huang CJ, Lin H, Yang XJ. Industrial Production of Recombinant Therapeutics in Escherichia coli and Its Recent Advancements. Ind. Microbiol. Biotechnol. 39(3) 2012: 383–399.
3 Aggarwal S. What’s Fueling the Biotech Engine: 2012 to 2013. Nat. Biotechnol. 32, 2014: 32–39; doi:10.1038/nbt.2794.
4 Bareither R, et al. Automated Disposable Small-Scale Bioreactor for High-Throughput Process Development: Implementation of the 24 Bioreactor Array. Pharma. Bioproc. 3(3) 2015: 185–197; doi:10.4155/pbp.14.64.
5 Tai M, et al. Efficient High-Throughput Biological Process Characterization: Definitive Screening Design with the ambr250 Bioreactor System. Biotechnol. Progr. 31(5) 2015: 1388–1395; doi:10.1002/btpr.2142.
6 Meusel W, et al. Recommendations for Process Engineering Characterization of Single-Use Bioreactors and Mixing Systems By Using Experimental Methods. DECHEMA Biotechnologie: Frankfurt am Main, Germany, February 2016.
7 Biener R, et al. Calorimetric Control for High Cell Density Cultivation of a Recombinant Escherichia coli Strain. J. Biotechnol. 146(1–2) 2010: 45–53; doi:10.1016/j.jbiotec.2010.01.004.
8 Luli GW, et al. Comparison of Growth, Acetate Production, and Acetate Inhibition of Escherichia coli Strains in Batch and Fed-Batch Fermentations. Applied Environ. Microbiol. 56(4) 1990: 1004–1011.
Corresponding author Dr. Marco Leupold (marco.leupold@sartorius-stedim.com) and Thomas Dreher are R&D scientists; and Dr. Gerhard Greller is R&D director of upstream technology at Sartorius Stedim Biotech in Goettingen, Germany; www.sartorius.com. Until recently, Mwai Ngibuini was ambr 250 product manager at Sartorius Stedim Biotech in Royston, UK. Contact his successor Barney Zoro at barney.zoro@sartorius-stedim.com.

