High–molecular-weight (HMW) and low–molecular-weight (LMW) product variants are critical quality attributes (CQAs) for monoclonal antibodies (MAbs) because they can cause severe immunogenic responses in human recipients. Aggregation is a common problem that can compromise the quality, efficacy, and safety of therapeutic proteins. It can occur at different stages in a biomanufacturing process: during cell-culture–based production, downstream process purification, drug-substance formulation, and storage of bulk drug substances or formulated drug products. Hence, the removal and control of MAb aggregates and fragments are crucial in bioprocessing.
Aggregation can be caused by covalent and noncovalent interactions in a protein molecule. Covalent aggregation happens when two or more monomers bind together or when unfolded and/or partially unfolded MAbs present binding sites to one another. Disulfide bonds can form between free thiol (free sulfhydryl) groups on MAb structures, another common cause of covalent aggregation. Noncovalent aggregates are formed by hydrophobic interactions that can occur between molecules that are very close together, such as in high-density cell cultures or highly concentrated formulations. Heterogeneity of protein content in solution can encourage protein aggregation. The most common cause of aggregation with the humanized MAb epratuzumab is the disulfide scrambling (1).
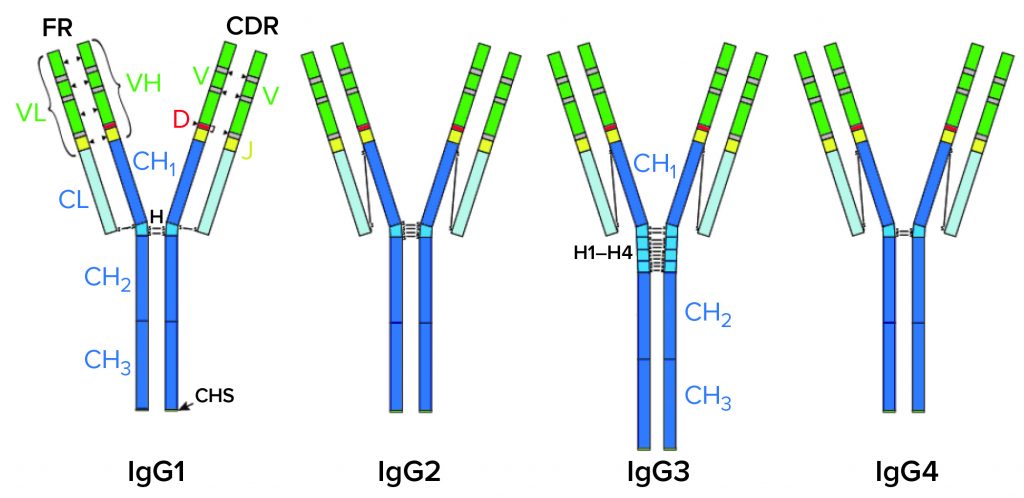
IgG Subclasses and Degradation Pathways: Immunoglobulins (IgGs) represent the most abundant class of antibody in human serum. Figure 1 depicts the four IgG subclasses. Recombinant IgG1-class antibodies can be more prone to aggregation and fragmentation than the others because of the extreme conditions they encounter in biomanufacturing processes (Figure 2). For example, aggregates can form during cell culture, making up ≤30% of the recombinant antibodies present in supernatant (2).
CH, CL = Constant heavy and light regions. VH, VL = Variable heavy and light regions. H = hinge region. CDR = Complementarity determining region. FR = Framework region. D = Diversity segment of variable chain.
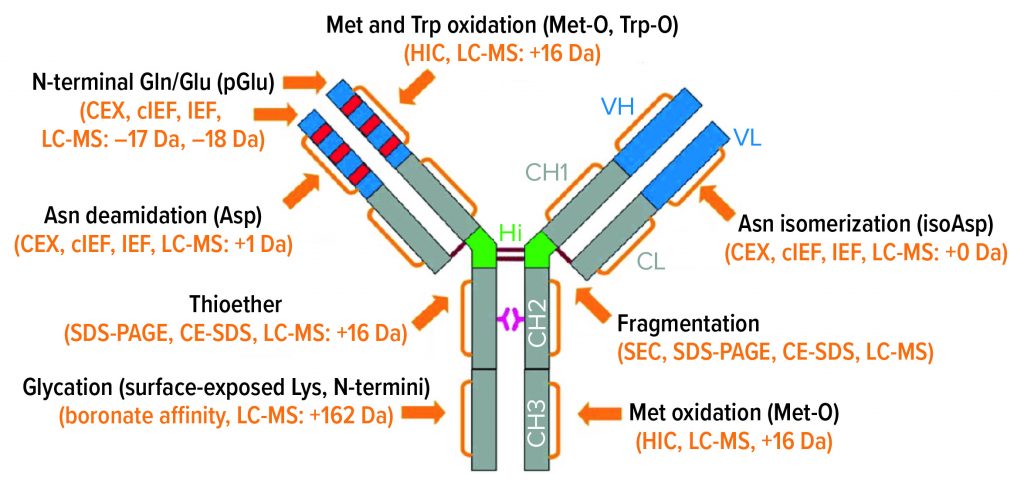
Figure 2: IgG1 structure and major degradation pathways, with analytical methods for investigating them. Definitions: CE-SDS = capillary electrophoresis with sodium dodecyl sulfate. CEX = cation-exchange (chromatography). cIEF = capillary isoelectric focusing. HIC = hydrophobic-interaction chromatography. IEF = isoelectric focusing. LC-MS = liquid chromatography with mass spectrometric detection. SDS-PAGE = sodium dodecyl sulfate polyacrylamide gel electrophoresis. SEC = size-exclusion chromatography.
During purification, low-pH elution conditions in affinity chromatography and agitation during mixing, pumping, and buffer exchange by ultrafiltration/diafiltration (UF/DF) all can cause aggregation of proteins in solution. Some buffers (such as polysorbates) and equipment contact materials (such as latex and single-use plastics) also have characteristics that can cause aggregation. And at the drug-product stage, similar effects of agitation, buffers, and product-contact surfaces can cause proteins to aggregate during formulation and fill–finish. Freezing, thawing, lyophilization, and reconstitution are particularly concerning because of pH changes, moisture migration, and other effects (3).
Objective, Methodology, and Framework
Bharat Serums and Vaccines Limited (BSV) in India studied the removal of aggregates (HMW) and fragments (LMW) from a MAb solution using affinity, cation-exchange (CEX), and mixed-mode chromatography (MMC) platforms. Our aim was to minimize the level of aggregates and fragments present to a minimum level without compromising overall MAb yield. We designed this study to measure the removal of aggregates from worst-case cell culture fluid (CCF) and worst-case downstream solutions to determine whether our process removes aggregates and fragments only from harvested supernatant or also from DS process intermediates.
For these experiments, we used a recombinant MAb in development at BSV and applied two primary analytical methods: size-exclusion high-performance liquid chromatography (SEC-HPLC) for aggregates and fragments and spectrophotometric analysis for step yields.
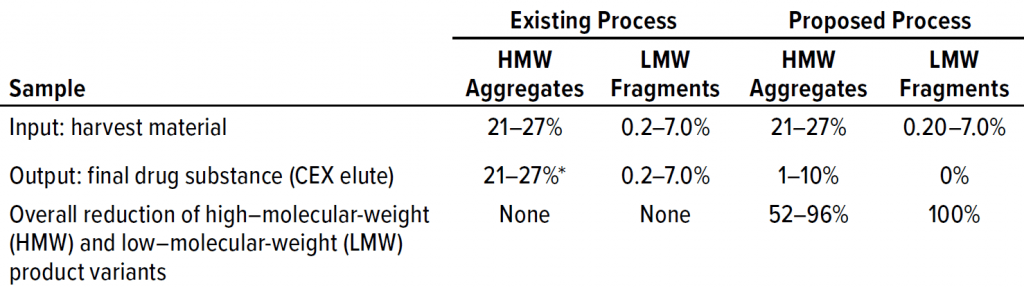
Our existing downstream process is incapable of removing aggregates appropriately. Product-variant levels after the final step have been ~30% HMW impurities and ~1% LMW impurities in the worst-case DS taken for study. For our proposed process, we tested two different approaches: one with harvested material processed using protein A affinity, multimodal, and cation-exchange chromatographic steps; and one in which the affinity step was skipped.
For the first process, 20 mL of MabSelect SuRe LX resin (Cytiva) was packed into a Cytiva XK 16/20 column at a bed height of 10 cm. We kept the operating flow rate at 4 mL/min considering a residence time of ≥5 minutes. In the intermediate step, we used Nuvia aPrime 4A (Bio-Rad Laboratories) mixed-mode resin and the same residence time. Bed height and volume were 2.5 cm and 5.0 mL, respectively in an XK 16/20 column. We kept the residence time to 5 minutes in flow-through mode. For a polishing step, we used a Capto-SP ImpRes ion-exchange resin (Cytiva) with similar residence time in bind–elute mode. These experiments ran on an ÄKTAprime plus chromatography system (Cytiva).
Capture Step: Dynamic binding capacity (DBC) of the protein A capture step was 58 mg/mL for different buffer conditions (Table 1), and elute was neutralized by a Tris-based buffer to pH 7.4.
Intermediate Step: Neutralized samples were subjected to MMC in flow-through mode with Nuvia aPrime 4A resin and a salt buffer (0.1 M glycine and 0.1 M NaCl) at pH 7.5 (4). We kept the DBC for this step between 40–50 mg/mL and calculated step yield by measuring MAb concentration with a spectrophotometer at OD280 nm. Aggregates/fragments contents were analyzed by SEC-HPLC.
Polishing: The final chromatography step was cation exchange with a strong cation resin as mentioned above and a column volume of 5.0 mL with 13–15 mg/mL DBC. Bed height was only 2.5 cm, although ideally more bed height is required for the best-resolution separations. In this case, we kept it minimal. Bed height is directly proportional to resolution (the higher, the better). But we have demonstrated that if our process works with a lower bed height, then it also will work with a higher bed height. Because of pressure issues with higher column beds, it is better to keep the lowest feasible height for manufacturing suitability.
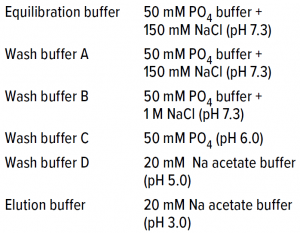
The buffer used for equilibration and wash was 20 mM sodium acetate at pHÂ 5.0; the elution buffer was 20 mM sodium acetate at pH 5.0 with 0.5 M NaCl at pH 5.5 for a 30-CV gradient elution. Load sample came directly from the Nuvia aPrime 4A flow-through fraction and loaded onto the is column after pH was adjusted to 5.0 with acetic acid. No conductivity adjustment was made, which provided an additional benefit of preventing the need for dilution while shortening the process time. We fractionated the elute and subjected it to response testing using SEC-HPLC and OD280 spectrophotometry.
Results and Discussion
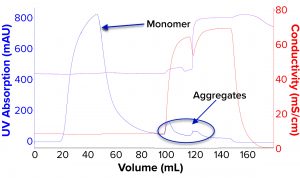
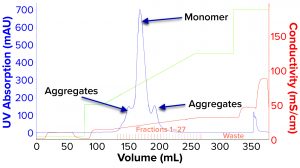
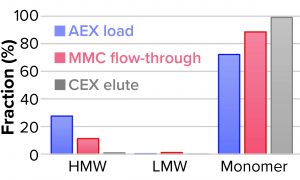
We evaluated removal of aggregates and antibody fragments from our first process run using MMC, and our results exceeded values reported by the manufacturer (Figure 3). Well-resolved separations yielded the monomer first, and aggregates later bound strongly to the resin. A repeat run with drug substance gave similar results for both the multimodal and cation-exchange resins (Figure 4). Again, separations were very good. Prepeak fragments (LMW variants) were separated by loosely binding with their less-positive charge, and the monomer formed a large middle peak. Aggregates (HMW variants) observed later were bound strongly by their greater number of positive charges.
Table 2 lists analytical results for aggregates and fragments from all runs. We observed reductions of aggregates (HMW%) and antibody fragments (LMW%) to ~5% and 2%, respectively, in the multimodal step — which indicates that MMC has worked very well. Figure 4 indicates reductions of aggregates and fragments alongside the monomer increments for each step in the new process. We used flow-through mode to minimize product loss. The new cation-exchange resin introduced in the proposed process also performed well in reducing HMW and LMW product variants. HMW reduction was to ~8%, and LMW fragments were removed completely. This gave a total removal for the process (Figure 5) of HMW to ~12% and LMW to 6.89% from harvest to CEX elution.

Table 2: Removal of high– and low–molecular-weight (HMW, LMW) variants during process runs against harvest taken from worst-case conditions; NA = not applicable.
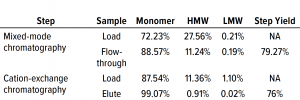
Table 3: Comparing high– and low molecular-weight (HMW, LMW) variant removal data from process runs against drug substance with large amounts of HMW and LMW present.
Table 3 lists reduction of HMW and LMW from drug substance as ~16% with MMC; no removal of LMW was observed here because the initial level was very low already. Figure 6 illustrates the same HMW and LMW reductions. Thus, our data indicate that the multimodal resin has done a good job in this process. Again, we used flow-through mode to minimize product loss. The cation-exchange resin also performed well with drug substance, providing a huge reduction of both HMW and LMW product variants — with complete removal of the latter. Total removal from drug substance was to ~28% for aggregates and to 0.21% for antibody fragments from drug substance to CEX elution.
Ongoing Process Development
Our existing process provided no reduction of HMW or LMW product-related impurities. Conversely, reduction of both was excellent in the proposed process (Table 4, Figures 7 and 8). We were pleased with the results for this particular IgG1 antibody captured by protein A affinity and purified by MMC and cation exchange. With multimodal resin in flow-through mode, it also aided in reduction of process-related impurities — for which data will be forthcoming from further studies.
The beauty of our new process was that it could remove product-related impurities at minimal bed height (2.5 cm only), considering that the height would be 10–15 cm for improved resolution. But the lower bed height improves manufacturability and eliminates high pressure concerns with the column, specifically for the smaller-bead cation-exchange resin used in this study. We are studying now other improvements in the proposed process to increase yield and reduce other types of impurities.
Note that our results apply only to the particular MAb product and conditions studied. You should not expect the same results without further optimization with your particular process and protein.
References
1 Remmele JR, et al. Active Dimer of Epratuzumab Provides Insight into the Complex Nature of Antibody Aggregate. J. Pharm. Sci. 95(1) 2006: 126–145; https://doi.org/10.1002/jps.20515.
2 Kramarczyk JF, Kelley BD, Coffman JL. High-Throughput Screening of Chromatographic Separations: II. Hydrophobic Interaction. Biotechnol. Bioeng. 100(4) 2008: 707–720; https://doi.org/10.1002/bit.21907.
3 Manning MC, et al. Stability of Protein Pharmaceuticals: An Update. Pharm. Res. 27(4) 2010: 544–575; https://doi.org/10.1007/s11095-009-0045-6.
4 Chen I, Vang L, He X. Poster: Nuvia aPrime 4A: A Hydrophobic Anion Exchange Resin for Refined Selectivity and Recovery. Bio-Rad Laboratories: Hercules, CA, 2020.
Further Reading
Frieden C. Protein Aggregation Processes: In Search of the Mechanism. Protein Sci. 16(11) 2007: 2334–2344; https://dx.doi.org/10.1110%2Fps.073164107.
Kameoka D, et al. Effect of Buffer Species on the Unfolding and the Aggregation of Humanized IgG. J. Biochem. 142(3) 2007: 383–391; http://dx.doi.org/10.1093/jb/mvm145.
Maggio E. Polysorbates, Immunogenicity, and the Totality of the Evidence. BioProcess Int. 10(10) 2012: 44–49; https://lne.e92.mwp.accessdomain.com/manufacturing/formulation/polysorbates-immunogenicity-and-the-totality-of-the-evidence-337120.
Patel J, et al. Stability Considerations for Biopharmaceuticals: Overview of Protein and Peptide Degradation Pathways. BioProcess Int. 9(1) 2011: 20–31; https://lne.e92.mwp.accessdomain.com/manufacturing/formulation/biopharmaceutical-product-stability-considerations-part-1.
Thorat AA, et al. Freezing-Induced Protein Aggregation: Role of pH Shift and Potential Mitigation Strategies. J. Controlled Rel. 323, 2020: 591–599; https://doi.org/10.1016/j.jconrel.2020.04.033.
Vidarsson G, Dekkers G, Rispens T. IgG Subclasses and Allotypes: From Structure to Effector Functions. Front. Immunol. 5(20) 2014: https://dx.doi.org/10.3389%2Ffimmu.2014.00520.
Wang W, Ignatius AA, Thakkar SV. Impact of Residual Impurities and Contaminants on Protein Stability. J. Pharmaceut. Sci. 103(5) 2014: 1315–1330; https://doi.org/10.1002/jps.23931.
Wang W, Nema S, Teagarden D. Protein Aggregation: Pathways and Influencing Factors. Int. J. Pharmaceut. 390(2) 2010: 89–99; https://doi.org/10.1016/j.ijpharm.2010.02.025.
Xiao N, et al. Leachables from Single-Use Systems and Impact to Product Quality. Single-use Technologies II: Bridging Polymer Science to Biotechnology Applications. Mahajan, Lye, Eibl-Schindler, Eds. ECI Symposium Series, May 2017; https://dc.engconfintl.org/biopoly_ii/43.
Corresponding author Vidya Kant Tiwari is assistant manager and downstream team leader, Pranav Gupte is manager, Mahesh Gavasane is general manager, Tushar Bhabal is executive, Saher Cheulkar is senior officer, and Saurabh Patwardhan is officer of biotech R&D in recombinant biotherapeutics downstream processing and formulation development at Bharat Serums and Vaccines Limited, Liberty Tower Third Floor, Airoli, Navi Mumbai – 400 708, India; 022-45043100; vidyakant.tiwari@bharatserums.com. ÄKTAprime, MabSelect SuRe, Capto, and Sepharose are registered trademarks of Cytiva (formerly GE Healthcare). Nuvia is a registered trademark of Bio-Rad Laboratories, Inc.