Immunoglobulin molecules are used extensively in therapeutic treatments, diagnostic applications, and fundamental academic research. Traditionally, full-length antibodies and smaller fragments such as the recombinant antigen-binding fragment (rFab) are produced through mammalian cell culture. rFabs also are small enough to be produced in Escherichia coli through fermentation (1, 2). Because disulfide bonds cannot be formed efficiently in the reducing cytoplasm of E. coli, rFabs are supplemented most commonly with a signal sequence that directs them to the more oxidizing bacterial periplasm for correct folding (3, 4).
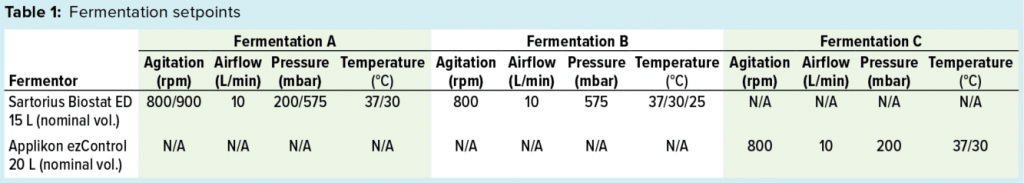
Figure 1a: Fermentation profile of a Sartorius Biostat ED system (15 L) using one temperature shift from 37 °C to 30 °C at the 6.5-h point; fermentation parameters that could be actively controlled during growth are shown on the right y-axes (pressure, airflow, agitation, and temperature), and parameters that were monitored during growth are shown on the left y-axes (dO2, pH, and acid/alkaline consumptions).
Relatively low yields present a major challenge in bacterial expression of correctly folded, functional rFab in E. coli. Most reported approaches to improve rFab yields in E. coli have focused on optimization of the expression construct, selection of media, and optimization of fermentation conditions, including aeration (5).
Dissolved oxygen (dO2) level has been reported to influence the secretion and production of Fab fragments (6). Below, we report that both the level of aeration and the control method of optimal dO2 have significant impacts on rFab yield. Three different fermentations are compared: Two use temperature control to take advantage of its effect on oxygen solubility and bacterial growth rate; the third uses direct enrichment with pure oxygen.
Harvested cell paste from each fermentation was lysed, then purified with Ni capture, anion-exchange (AEX) chromatography, and diafiltration. Dramatic differences in yield were observed because of the strategies used to control dO2 during fermentation. Although more research is needed to understand how such approaches differentiate yields, our work highlights the importance of developing rFab production processes based on fermentation.

Figure 1b: Fermentation profile of a Sartorius Biostat ED system (15 L) using a double temperature shift from 37 °C to 30 °C at 5.5 h and to 25 °C at 9.4 h; fermentation parameters that could be actively controlled during growth are on the right y-axes (pressure, airflow, agitation, and temperature), and parameters that were monitored during growth are on the left y-axes (dO2, pH, and acid/alkaline consumptions).
Materials and Methods
Bacterial Strain: We used Abbott-sourced DH10B E. coli cells with recombinant plasmids controlling expression of target rFab with both a heavy and light chain. Expression of the target rFab was induced by adding sterile isopropyl-β-d-thiogalactopyranoside (IPTG, Sigma I6758) and arabinose (Sigma A3256-500G) solutions at final concentrations of 1.0 mM and 12.0 mM, respectively.
Composition of Complex Medium: Composition and preparation of batch and feed media are proprietary to Abbott. The feed medium contains carbon source only. After preparation and sterilization of that medium either by autoclave or sterilization in place (SIP), sterile tetracycline (Sigma T7660) and chloramphenicol (Sigma C0378) were added before inoculation at final concentrations of 10 g/mL and 50 g/mL, respectively.
Cultivation — Seed Culture Growth: A 500-mL culture in a 2.5-L sterile Thompson disposable polycarbonate Erlenmeyer flask grew overnight to an optical density at 600 nm (OD600) range of 12.0 ± 4.0, then was used to inoculate the bioreactor for batch cultivation. The culture was stored at 4–8 °C until inoculation, which happened within 24 hours of preparation.
Cultivation — Fermentation: We carried out fed-batch, high–cell-density fermentations in Applikon ezControl (20 L) and Sartorius Biostat ED (15 L) stainless-steel fermentors. Both systems have SIP for sterilization and cleaning, standard sensors for pH and dO2 control, and exhaust O2 and CO2 monitoring with a BlueInOne off-gas sensor from BlueSens. Only the Applikon system was equipped with a module for pure O2 enrichment.
Before starting each fermentation, we calibrated the dO2 sensor to 100% at identical settings of pressure (200 mbar), airflow (10 slpm), agitation (800 rpm), and temperature (37 °C). The fermentor was started at a temperature of 37 °C, a pressure of 200 mbar, gas flow rate of 1.0 vvm, and pH control at 6.8 with a dead band of 0.1. We used the exponentially grown starter culture to inoculate the fermentor to OD600 of 0.8–1.0, with a working volume of 8.0 L. The feed started when OD600 exceeded 1.0, typically within three hours of inoculation, with a constant rate of 3.0 mL/L/h. For induction of rFab expression, we added IPTG and arabinose when OD600 reached a target of ~50.0 ± 4.0. After induction, fermentation was extended for 12–15 hours and harvested when OD600 reached a target of ~100.0 ± 10.0.
To control dO2 above the setpoint of 30% or to at least minimize oxygen depletion, we implemented different strategies based on the capabilities of each fermentor system. Table 1 lists exact setpoints for each fermentation.
Purification: The entire cell paste from each fermentation in this study was suspended in 2 L of lysis buffer and lysed by continuous sonication for one hour at 80 A. After centrifugation and filtration, soluble rFab was purified using cOmplete His-tag resin (from Roche) followed by AEX chromatography (Q HP columns from Cytiva) in flow-through mode. We performed diafiltration to place the rFab to a final storage buffer.
Purification and final formulation details are proprietary to Abbott.
Analytical Method
During cultivation, we took 10-mL samples at multiple timepoints and determined their OD600 values. For harvest, the entire fermentation broth was centrifuged at 13,000 rpm for 25 minutes using a Beckman J2-21 centrifuge with JLA8.1 rotor. After decanting the supernatant, we calculated pellet weight as wet weight biomass growth and passed the cell paste through purification. We evaluated purified rFab using A280 analysis, size-exclusion chromatography high-performance liquid chromatography (SEC-HPLC), mass spectroscopy (MS), and sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). Results showed purity >98% (data not shown).
Results and Discussion
Sartorius Biostat Fermentation Using a Single Temperature Shift for dO2 Control: We performed one temperature shift from 37 °C to 30 °C at the 6.5-h timepoint (Figure 1a) when dO2 dropped to ~10%. At that time, OD600 was 23.2. As we dropped the temperature, pressure elevated from 200 mbar to 575 mbar, and we kept both settings through the remainder of the fermentation. Figure 1a shows the fermentation profile for this growth.
Fermentation parameters actively controlled during growth were pressure, airflow, agitation, and temperature. Parameters monitored during growth were dO2, pH, and acid and alkaline consumption. We observed that dO2 increased from its lowest level of ~10% (after changes in temperature and pressure). That value dropped again following induction of expression but remained higher than ~18%. We also observed a pH decrease throughout the fermentation as carbohydrate and other nutrients were consumed. We controlled pH at a setpoint of 6.8 through constant base consumption.
We induced the fermentation by adding IPTG and arabinose solutions to final concentrations of 1.0 mM and 12.3 mM, respectively, after about nine hours at OD600 = 48.8. At the time of induction, we increased agitation from 800 rpm to 900 rpm, where it remained until fermentation ended. We harvested the fermentation after 21.5 h of growth and at OD600 = 91.5. Yield was 1057.5 g of cell paste, which was purified entirely to yield 60 mg of rFab.
Sartorius Biostat Fermentation Using a Double Temperature Shift for dO2 Control: We began this fermentation by increasing the pressure at fermentor inoculation from 200 mbar to 575 mbar, which was maintained throughout the growth. The first temperature shift, a decrease from 37 °C to 30 °C, occurred at 5.5 h when the dO2 dropped to ~30%. Following a brief rebound, dO2 dropped again to ~30%, at which time we adjusted the temperature down to 25 °C (Figure 1b).
At the second temperature adjustment, we measured OD600 = 46.8 and induced expression by adding IPTG and arabinose at the same levels used in the single-temperature–shift experiment. We controlled dO2 to >30% throughout the fermentation and harvested after 25 h of growth at an OD600 = 97.6. Yield was 1272.0 g of cell paste, which was purified entirely to yield 250 mg of rFab.
Fermentation Using One Temperature Shift and Pure Oxygen Enrichment: We performed the third fermentation using an Applikon ezControl fermentor (20 L), which was equipped with a pure-oxygen enrichment module. To ensure that dO2 levels could be compared across both fermentor brands, we calibrated dO2 to 100% at the same levels of agitation, temperature, airflow, and pressure. The parameters used in the Applikon-fermentor run were identical to those used in both Sartorius-fermentor studies except that we activated a cascade control of dO2 at 30% using pure oxygen enrichment. We initiated pure oxygen supplementation at five hours and 17 minutes, when dO2 fell close to the 30% setpoint (Figure 2). In the first few hours of control, dO2 levels bounced between ~15% and above 100%, stabilizing at the 30% setpoint at 10.5 h of growth.

Figure 2: Fermentation profile of an Applikon ezControl system (20 L) using one temperature shift and pure oxygen enrichment; pure oxygen supplementation initiated at 5 h and 17 min. Temperature shift (37 °C to 30 °C) was initiated at induction at 8.5 h of growth. Fermentation parameters that could be actively controlled during growth are shown on the right y-axes (O2 enrichment, pressure, airflow, agitation, and temperature), and parameters that were monitored during growth are shown on the left y axes (dO2, pH, and O2/CO2 concentrations).
At the 8.5-h timepoint, we measured OD600 = 46.6 and induced expression using the same levels of IPTG and arabinose that we used in the previous runs. At induction, we reduced temperature from 37 °C to 30 °C and held it constant for the remainder of the run. We harvested the fermentation at 16 h postinduction and at OD600 = 90.8. Yield was 1171.1 g of cell paste, which was purified entirely to yield 1357 mg of rFab.
Discussion
Controlling dissolved oxygen and keeping it above a certain level is crucial in many bioproduction processes. That level depends on a given clone and process. We observed three different dO2 profiles (Figure 3a). The single temperature shift resulted in a dO2 level running below the levels in the double–temperature-shift study and pure-oxygen–supplemented study and lower than the targeted level of >30% during the induction phase of growth. The dO2 level of the double temperature shift ran above the 30% setpoint postinduction. Differences in the biomass growth curves (Figure 3b) and final cell paste yields (Figure 3c) for the three runs were insignificant.
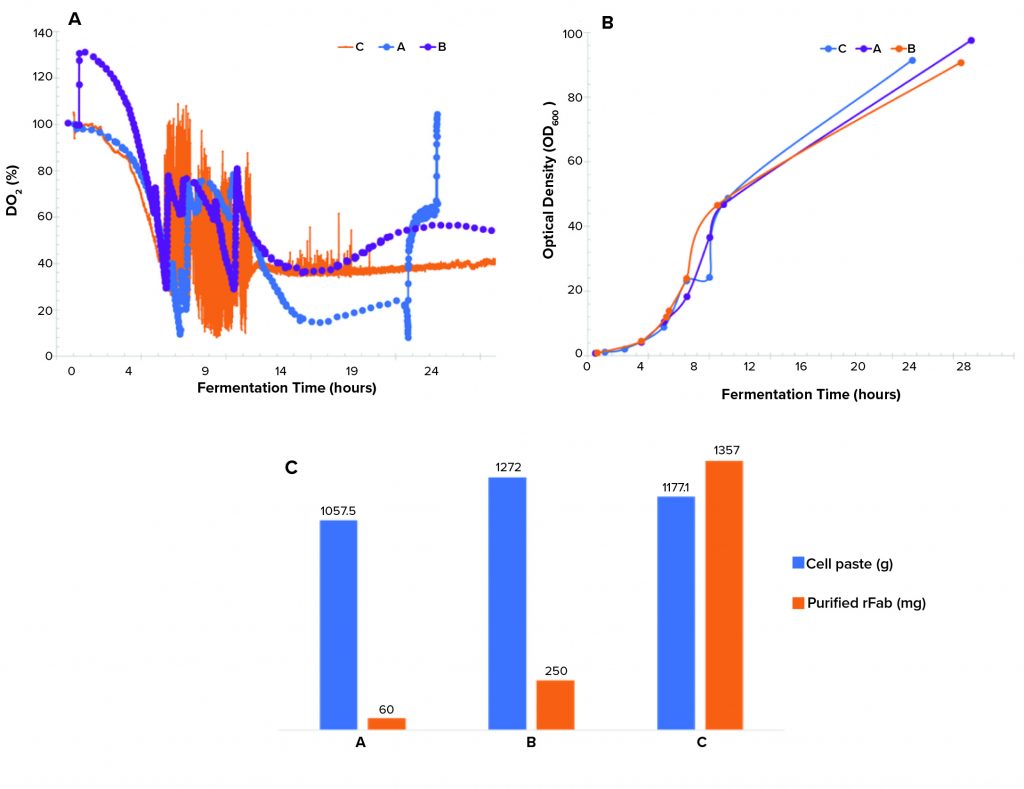
Figure 3: (a) Dissolved oxygen profiles, (b) biomass growth curves by OD600 measurement, and (c) cell paste and purified rFab yields across three fermentations; (a) is the Sartorius fermentor, single temperature drop; (b) is the Sartorius fermentor, double temperature drop; and (c) is the Applikon fermentor, single temperature drop and O2 supplementation.
A point of interest is the observation that purified rFab yields were significantly different for the three fermentation runs (Figure 3c). Those data indicate that the way by which dO2 is controlled (through a cascade control at 30% postinduction) directly influenced the level of rFab expression obtained in the Applikon fermentation process. The methods demonstrated in our studies of dO2 control are standard strategies applied during fermentation process development.
For the runs performed with a Sartorius fermentor, we used a different temperature-reduction strategy. That influenced oxygen solubility to sustain oxygen demand that was required for the high-density growth (because this system could not provide a pure-oxygen supplement). Temperature reduction also could be linked to a decrease in rFab production in the Sartorius-system runs. That is because temperature changes affect not only oxygen solubility, but also numerous metabolic and biologic activities (7, 8). It is possible that the flux observed in dO2 in the Sartorius fermentations because of the temperature changes influenced the redox environment of the periplasmic space, where production of functional fab fragments takes place, resulting in lower expression of rFab in that study. That theory is supported by the higher yield of rFab obtained from the Applikon fermentation. In that case, we maintained a constant dO2 level postinduction. Our continuing studies are focused on determining the mechanism of how dO2 control is affecting rFab yield.
References
1 Pack P, et al. Improved Bivalent Miniantibodies, with Identical Avidity as Whole Antibodies, Produced by High Cell Density Fermentation of Escherichia coli. Nat. Biotechnol. 11(11) 1993: 1271–1277; https://doi.org/10.1038/nbt1193-1271.
2 Francisco JA, et al. Production and Fluorescence-Activated Cell Sorting of Escherichia coli Expressing a Functional Antibody Fragment on the External Surface. Proc. Natl. Acad. Sci. USA 90(2) 1993: 10444–10448; https://doi.org/10.1073/pnas.90.22.10444.
3 Brecht B, et al. Cytoplasmic Versus Periplasmic Expression of Site Specifically and Bioorthogonally Functionalized Nanobodies Using Expressed Protein Ligation. Protein Express. Purif. 133, 2017: 25–34; https://doi.org/10.1016/j.pep.2017.02.009.
4 Schatz G, Dobberstein B. Common Principles of Protein Translocation Across Membranes. Science 271(5255) 1996: 1519–1526; https://doi.org/10.1126/science.271.5255.1519.
5 Kaisa U, et al. Effect of Culture Medium, Host Strain, and Oxygen Transfer on Recombinant Fab Antibody Fragment Yield and Leakage to Medium in Shaken E. coli Cultures. Microb. Cell Fact. 12, 2013: 73; https://doi.org/10.1186/1475-2859-12-73.
6 Konz JO, King J, Cooney CL. Effects of Oxygen on Recombinant Protein Expression. Biotechnol. Prog. 14(3) 1998: 393–409; https://doi.org/10.1021/bp980021l.
7 Sørensen HP, Mortensen KK. Soluble Expression of Recombinant Proteins in the Cytoplasm of Escherichia coli. Microb. Cell Factories 4(1) 20015: 1; https://doi.org/10.1186/1475-2859-4-1.
8 Schein CH. Production of Soluble Recombinant Proteins in Bacteria. Biotechnol. 7, 1989: 1141–1148; https://doi.org/10.1038/nbt1189-1141.
Corresponding author Yongxue Ding is principal scientist, You Pan is principal research scientist, Mark Gibson is group leader of the propagation team, Troy McSherry is leader of the purification group. and Steven P. Allen is a Volwiler Research Fellow and manager of biologics process design and analytical chemistry research and development, all at Abbott Diagnostics Division, Department 09NC, AP8A, Abbott Laboratories, 100 Abbott Park Road, Abbott Park, IL 60064; 1-224-668-3629; yongxue.ding@abbott.com.