Artificial intelligence (AI) in healthcare and biopharmaceutical industries still is in early stages of development and use, yet proper data management is critical to successful implementation at any stage of the enterprise continuum. AI is applied to help researchers understand and analyze biology, diagnose diseases, design new drugs, and predict clinical potential and treatment outcomes. AI has been applied in the discovery and development of novel drugs and for repurposing existing drugs, manufacturing different types of drug products, and optimizing biocommodity production. It has become an essential tool in assessing real-world evidence and for data processing using “-omics,” next-generation sequencing, clinical trial design, and personalized patient management. AI also is used in clinical applications for maintaining patient electronic health records (EHRs), population health management, predictive care guidance, digital imaging, digital pathology, and diagnostics.
Healthcare organizations should aim to educate themselves about AI, including its benefits and its limitations, and to think about the opportunities this umbrella of technologies can make possible. Healthcare organizations also can adopt AI-based solutions or choose to develop or codevelop their own. That path requires commitment and investment — understanding AI best practices and the systematization of AI-based approaches in data capture, data curation, and agile implementation strategies according to proven methodologies.
Below, we overview the different components and considerations of big data (including the different types of data) and AI applications in bioprocessing and healthcare settings.
Data Types
Raw Data: When someone mentions the word data, typically a data table comes to mind, perhaps as a spreadsheet with column names along the top and row after row of numbers or text down the length of the table. Although many other types of data sets exist, we start with this type of presentation as a basic example. The first consideration is the type of data in each cell of the data table. The two broad categories are structured and unstructured.
Structured data types include the following:
• Scalar data are single elements that describe a magnitude (e.g., the number –13.7).
• Vector data are one-dimensional lists of elements of a fixed length (e.g., the bitvector 10010101110).
• Array data typically refer to two-dimensional tables of data (e.g., a matrix).
• Tensor data are of three or more dimensions and include complex matrices of data consisting of individual elements of any data type.
Unstructured data types include the following:
• String data typically are text characters and can be of any length (e.g., ABCDEFG).
• Image data include photos and cross-sectional digital pathology slides.
• Video data include, for example, time-lapse animations of a virus infecting a cell.
• Audio data include digital recordings of patients coughing from a respiratory tract infection.
Data set size (e.g., the number of rows and columns in a table) is an important consideration. A data set must be large enough to allow for the proper training of machine-learning models. Table 1 shows a portion of a data set available from a US Food and Drug Administration (FDA) website.
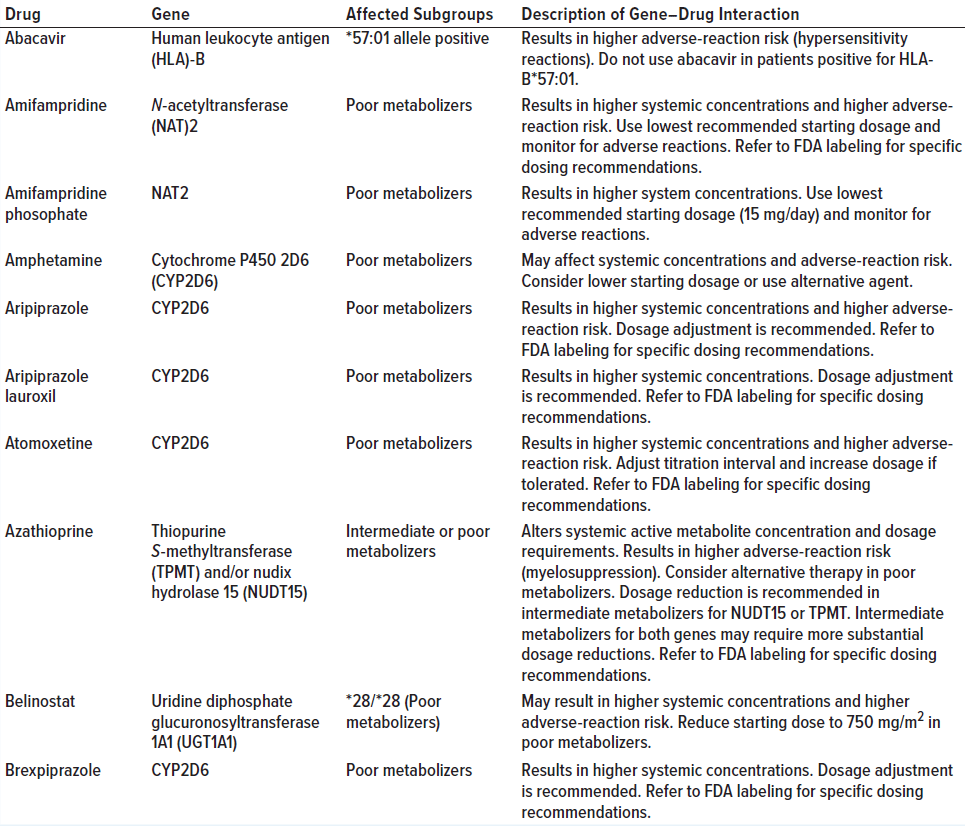
Table 1: Example of a data set (pharmacogenetic associations) available from an FDA website (adapted from https://www.fda.gov/medical-devices/precision-medicine/table-pharmacogenetic-associations); the “dirty data” section on page 5 discusses data quality using the entries in this table as examples.
Internal and External Data Sets: Healthcare organizations routinely generate large volumes of data as a result of their regular operations. These include data related to patient characteristics, personal records, medication details, financial transactions, and so on. That information serves as a rich source of internal data that can be extracted from internal data management systems. Some external data sources are publicly available data sources that can be used to gain further insights.
Data ingestion is defined by the transportation of data from diversified sources to a storage medium in which those data become accessible, usable, and analyzable by a company or organization. Having the right data ingestion mechanism is critical as data volumes increase both in quantity and source variation. Within healthcare and bioprocessing, data are gathered from multiple sources and applications, including medical devices, wearables, internet of things (IoT) devices, patient-reported outcomes (PROs), EHRs, diagnostics, imaging, enrollment records, claims data, and billing. In almost all enterprises, data silos exist through those applications, which can lead to structured, semistructured, or unstructured data at different “velocities” and in different volumes. With diverse and complex data formats, structure, and context, gathering, processing, and harnessing key information into useful, actionable assets can be difficult. Recent developments in big data integration can be used to help companies and organizations manage data silos efficiently, thus improving data mapping and accuracy.
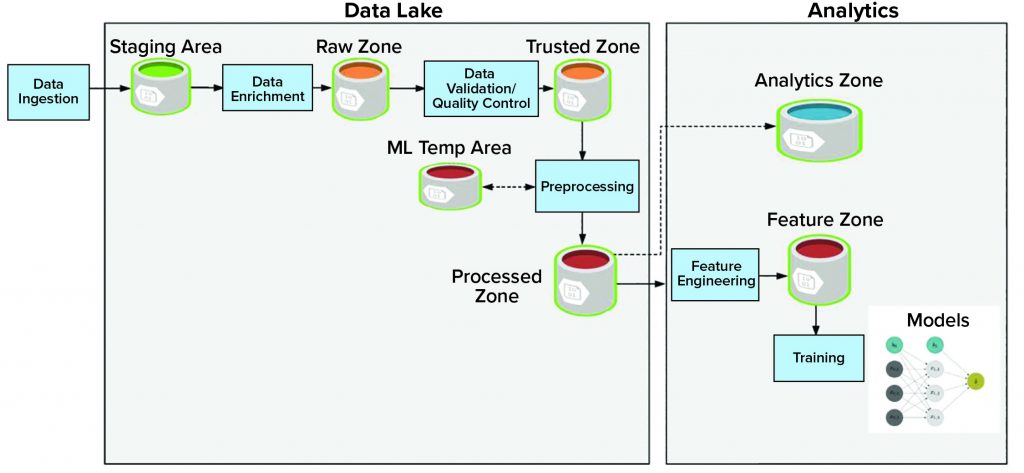
Storage: A data lake or data repository stores vast amounts of data and holds both structured and unstructured data. The data lake consists of multiple data zones (Figure 1):
• The staging zone contains source data from data producers in its original format. This area is a temporary storage area.
• The raw zone contains all raw data after initial data enrichment.
• The trusted zone is the source of trusted data after performing data quality and validation.
• The processed zone stores output results after data processing jobs are executed.
• The machine-learning temporary area stores temporary data generated by different preprocessing and machine-learning data jobs. This area provides high input and output throughout.
Adjacent to the data lake is analytics storage, which itself houses different components of the data workflow and consists of different zones:
• The feature zone is the central place to store curated features for the machine-learning pipeline.
• The analytics zone is designed to perform advanced analytics and complex queries.
Data federation technology enables the creation of a physically decentralized but functionally unified database, collecting and storing metadata rather than actual data. Constituent databases are interconnected virtually and can be geographically disparate. That is an alternative approach to creating centralized data sources because constituent database systems remain autonomous and reduce the amount of effort required for merging several disparate databases. The primary use of data federation is to access and merge already cleaned and conformed data in real time. That allows for different sources of data (within a company or organization) to keep data within established firewalls.
| Transformations in Biomanufacturing Through AI-Enabled Systems
The increasing amount of digitization of in our world today introduces astounding new discoveries and challenging new questions. Whether it be in the modes of human connection, accessibility of information, or the interoperability of systems across all phases of our daily lives, the 21st century is centered on the collection, interrogation, and exploitation of data-driven systems. The current transformation perhaps is the longest running and most common to all forms of manufacturing. The way in which we think of new concepts, test their validity, and enact them at a macroscale is undergoing a watershed transformation. Since the late 1970s, traditional concepts in the design, development, and execution of material production have been slowly transformed by the increasing introduction of computer-based, data-driven decision-making into our workflows. As more data have been leveraged in improving efficiencies across the different phases of manufacturing, innovators have fed the insatiable demand for better, faster, and more effective solutions to augment the capabilities of humans in different processes. That trend has led to improved delivery of services and control over costs, resulting in the adoption of data-driven practices across all manufacturing sectors. In recent years, this transformation has been captured under the catch-all term of artificial intelligence, useful in describing the myriad systems that have morphed from the realm of science fiction into necessary daily companions. The adoption and implementation of AI — and more specifically, machine learning — into manufacturing workstreams has had and will continue to have a tremendous influence on both producers and consumers across different industries. Most recently, AI and machine learning have been leveraged in healthcare – a seemingly discordant delay in adoption given the wealth of data generated, the complexity of the questions posed, and the critical necessity for improvements in the delivery of care. However, despite this lag in adoption, AI and machine-learning solutions have become the cornerstone of the future of medicine, influencing the entire spectrum of healthcare. AI and machine learning also have found a welcome home in discovery, development, validation, diagnostics, patient selection, and delivery of care. Parallel and upstream to such applications is the incipient transformation in biomanufacturing, which in many ways mirrors and in many others expands beyond the strides made in trying to improve “the patient experience.” Biomanufacturing driven by AI and machine learning, also described as intelligent manufacturing, enables a transformative union of stakeholders in therapeutic discovery and development. A traditional approach to biomanufacturing is predicated on years of experience, reams of data, and the identification of critical quality attributes, around which the success and failure of bioprocessing are based. In bioprocessing, a therapeutic program progresses from discovery to research and development (R&D) and through process development before entering a defined chemistry, manufacturing, and control (CMC) program that enables product validation in clinical trials. During each stage of bioprocessing, what biomanufacturers are looking for in a product becomes increasingly well defined. Often, however, “missed opportunities” in early data are poorly interrogated because of constraints in data capture and available analytical systems. Aspects dictating the performance of a program can be identifiable from trends in early data only if the tools for objective assessment are leveraged. Such innovations (in both the time-saving solutions in data architecture and in the dynamic capabilities of an algorithm to adapt to unusual and unstructured data) will result in shorter timelines and lower costs of development in even complex biologics-based therapeutic programs. On the other side of the spectrum, once a drug candidate has entered clinical validation, the findings from those trials often are challenging to correlate to early data sources and are underutilized in iterative product development. The introduction of more rational data capture and data analytics engines to the assessment of clinical path programs offers a unique opportunity to learn more about a drug product and to better guide the development of its next-generation candidates. Feeding data from clinical trials back into the development pipeline provides a tremendous opportunity for rationale product improvement. Perhaps in the future, analytics will be sufficient enough to enable small-scale, precise clinical trials performed not only to test the efficacy of a current product, but also to allow for refinement of the product for further use. The future opportunities afforded by the introduction of AI and machine-learning platforms to biotechnology, in particular biomanufacturing, are incredible. Integration of rationale data-capture solutions coupled with unbiased analytical systems in R&D and manufacturing will allow for faster development of less expensive drug products better suited to target patient populations. |
Working with Data
Quality Control: Another important attribute is data quality. Data are the most crucial asset for all AI undertakings, but those data are of no value if they are unreliable. Machine learning is the process of using statistical techniques on data and training computers to operate on their own. Thus, data quality plays a significant role because machine learning algorithms learn from those data.
When considering data quality, it is important to determine whether the source of those data was meant for a human reader or for software programs. Because sources and formats of data vary, ensuring data quality is important. The use of large data sources, streaming data, complex data, and unstructured data can complicate quality issues as well. Data quality always has been an issue with database and data collection systems. However, the types of data errors and their potential consequences are different when used in AI and machine learning. In AI, models and data quality are intrinsically linked using training data. Models and algorithms can be thought of as scientific experiments: If the wrong data are selected, then an experiment can fail to produce an adequate result. The need for large data sets for training models further complicates problems because of a high risk of common data issues such as noise, missing values, outliers, lack of balance in distribution, inconsistency, redundancy, heterogeneity, data duplication, and integration.
Dirty Data: Poor quality can create the most significant bottlenecks in data science. Table 1 shows example of the common types of poor data quality (dirty data):
• Incomplete — not all items listed under the “Affected Subgroups” heading contain information about the allele.
• Duplicate — the left-hand column lists amifampridine twice (albeit with a salt form the second time).
• Inaccurate or incorrect — several of the rows of the “Affected Subgroups” table have a “*” or “/” in the description. This inconsistency will trip up automatic parsers.
• Inconsistent — the first gene has a hyphen in the name, but the others do not.
• Outdated — many drug names have not been updated in this version of the table.
Data preprocessing is a data-mining technique that involves transforming raw data into an understandable format. Raw data refers to the data in its source form, without any prior preparation for performing analytics or machine-learning operations. Data preprocessing is a necessary and integral step in AI workflows. For example, healthcare data often are incomplete, inconsistent, and short on behavior trends (e.g., lacking attribute values and certain attributes of interest). Such data often contain only aggregate data, errors or outliers, or discrepancies in codes or names. The quality of data and the useful information that can be derived from them directly affect the ability of a model to learn. Thus, preprocessing is an important step before such data can be used for AI.
Data preprocessing includes operations such as data cleansing, curation, integration, reduction, entity resolution, and data transformation. For data used in AI applications, preprocessing also includes partitioning data points from input data sets to create training, evaluation (validation), and test sets. That process includes techniques for repeatable random sampling, minority-classes oversampling, and stratified partitioning.
In its simplest form, data aggregation organizes data into a comprehensive and consumable format (to provide analysts with insights). Because of the exponential growth of data collection and its increasing accessibility, the application of data aggregation has become commonplace and necessary. Data aggregation is industry agnostic and often critical to the success and continuous improvement of organizational operations across the world. However, it has become especially relevant to the healthcare industry, where the benefits of aggregation are numerous and can lead to better insights for both healthcare providers and patients. With advances in computing and technologies such as AI and machine learning, the scale and capacity of data aggregation have grown. In the AI and machine-learning environments, aggregation is performed at the data and model levels. Data are preprocessed to generate an aggregate that yields improved results from the same data. Aggregated models tend to yield results that are more accurate than individual models.
Visualization and Exploration: Using visualization tools such as charts and graphs — whether static or as interactive dashboards — AI users and domain experts can explore data in different ways. Doing so can help analysts detect weird trends, outliers, and other data anomalies. This phase often is aligned with quality control because visualizations can provide some insights during the inspection of raw data, which in turn can help analysts ask the right questions. Thus, companies can use visualization to determine the steps to take during a feature-engineering phase. Later in an AI process, visualizations can help stakeholders explore the learnings of trained models, the contributions of features to model accuracy, and other useful explanatory aspects of the model.
Feature Engineering, Extraction/Selection: In machine learning, the way in which data are represented plays a pivotal role and often is the difference between a good or bad model. Feature engineering is the process of creating a “good representation” of the data to be modeled. The term good representation is subjective and depends on the problem to be solved and the goal of a specific modeling project. For example, in healthcare, one of the most important aspects is interpretability to clinicians. Thus, interpretability should be an important consideration while creating “good representations.”
Generally, feature engineering is an iterative process. A person performs an iteration of feature engineering and modeling, notes the model performance, and continues to iterate until the desired performance is reached. Traditionally, feature engineering involves domain experts handcrafting features according to their experience, but recently, there has been a trend to use deep-learning algorithms to perform feature engineering and end-to-end modeling.
Data Pipelines: The multiple steps of data collection and preprocessing typically are sequential. As such, those steps are best constructed using a pipeline concept. After one task finishes, the next in the series begins. Once designed and debugged, that workflow of tasks can be automated and set to run on a predetermined schedule. If new input data are the same type and format as the existing data repository, then automation can speed the process and eliminate the need for user intervention.
That process can become much more complicated when new data to be entered into a system are of different types and formats. In such cases, it is important to address common pitfalls. Two of the most haphazard approaches to accommodating different data are to amend directly the underlying database that stores processed feature data and to tweak systems to allow for new data. If that is done without consideration for the automation workflow that initially was designed to generate that system, however, the pipeline might be essentially “broken” and cannot be used in the same way again. That problem worsens when different tasks or scripts of the original pipeline are patched to further accommodate different input data — perhaps by new developers and system architects that were not involved in development of the initial workflow. Although that approach can be tempting as a quick fix, it can be a knock-on effect of long-ranging consequences. For example, because the patch was created to remedy a specific task in the pipeline, it is unknown whether automation of the entire pipeline will lead to desired results. Best practices for such a scenario are to destroy or archive the original database and systems and rebuild them using an updated automation workflow.
Processed Data: Following data cleaning and feature extraction, the total data available then are divided into three subsets: training, validation, and test data (described in the next section). The reason for that division is so that a model can be developed on one subset of the total data (training set), then the performance of multiple models can be validated (validation set) so that the best candidate is chosen. Finally, testing (test set) is performed to gain a good understanding of what the real-world performance of the model is likely to be.
Before splitting a data set into those three subsets, it is important first to understand the origins of the data and the sources for different components. For example, sections of data could come from measurements at different points in time (days, months, years) or different subpopulations split by age or geography. Those natural subsections of the total dataset must be represented adequately in all the training, validation, and testing sets.
Note that determining how real-world data (to which a model will be applied) might be different from the original data set is important. A model that has been trained on data that are significantly different (e.g., because of a drift in health of a population over time) might be inaccurate.
Building Models with Data
Among the three data subsets described previously, the training data set (or reference data set) typically is the largest. It should be a large enough sample with sufficient variability across the total set so that it can be used to train a model’s parameters and hyperparameters accurately. Training data also are used for data exploration and analysis, which is part of choosing a model correctly.
During model development and training, a validation data set is used to monitor the performance of parameter “tuning.” Each time a new parameter set is developed on a training data set, its performance is validated from the validation data set.
A test data set (or query data set) is one that is used solely to indicate real-world performance to be expected of a model. Those data should never be used to train parameters.
Metadata is used to describe actual data for consistent and predefined cataloging of data, without the need to go through or process actual data. Metadata comes in different formats. Below, we distinguish three different types: business, technical, and operational metadata.
Business metadata often is used to describe external metadata because it does not describe the content of data but rather how those data are being used, from where they originate, where they are stored, and how they relate to other data that are being stored in a data warehouse.
Technical (internal) metadata is used to describe actual data. Technical metadata defines the objects, data structures, data types, fields, and so on. It is used to develop data processing routines and to determine the type of data that can be expected from that processing.
Operational metadata is used to describe the processes and events that take place when data are being processed. Examples include the name and status of a data-processing job, the computer (system) on which the processing took place, and so on. Operational data should be descriptive and complete enough that when reprocessing is performed, the same results are achieved. In that way, analysts can trace how data ended up in a certain state.
Keys for Implementation
The future impact of AI in healthcare and biological manufacturing will depend on the broader community’s commitment to best practices around data capture and quality. Data standards will play a key role. Many competing standards exist, and although it may be tempting for organizations to invent their own frameworks, it may be more beneficial to adopt and consolidate existing systems. Likewise, standards for algorithm development and performance will be essential, and those should come from collaborations among AI practitioners, regulatory agencies, and industry and patient advocates. In the healthcare industry, for example, such work is underway in conversations between the Alliance for Artificial Intelligence in Healthcare (AAIH) and the FDA.
Considerable investments in education at all levels — including for executives, developers, technicians, healthcare providers, and patients — are warranted to ensure that AI is used where it can make a difference and that solutions are developed that subscribe to appropriate methods and guidelines. Successful implementations of AI for improving healthcare outcomes and bioprocessing efficiencies should be championed. But companies and organizations shouldn’t run from failure either. Not everything will work, and learning from missteps rather than discounting an entire endeavor will be key. We can draw an analogy from the design–build–test paradigm familiar to the synthetic biology and bioprocessing worlds. Or perhaps the right analogy comes from AI itself: The entire healthcare industry will learn and improve its accuracy with the more data it observes.
Further Reading
Moore C. Harnessing the Power of Big Data to Improve Drug R&D. BioProcess Int. 14(10) 2016; https://lne.e92.mwp.accessdomain.com/manufacturing/information-technology/harnessing-power-big-data-improve-drug-rd.
Montgomery SA, Graham LJ. Big Biotech Data: Implementing Large-Scale Data Processing and Analysis for Bioprocessing BPI. BioProcess Int. 16(9) 2018; https://lne.e92.mwp.accessdomain.com/manufacturing.information-technology/big-biotech-data-implementing-large-scale-data-processing-and-analysis-for-bioprocessing.
Richelle A, von Stosch M. From Big Data to Precise Understanding: The Quest for Meaningful Information. BioProcess Int. 18(2) 2020; https://lne.e92.mwp.accessdomain.com/manufacturing/information-technology/systems-biology-tools-for-big-data-in-the-biopharmaceutical-industry.
Braatz R. New Data Analytics Tools Can Improve Bioprocess Workflows — If Applied Correctly. BioProcess Int. 18(9) 2020; https://lne.e92.mwp.accessdomain.com/manufacturing/information-technology/new-data-analytics-tools-can-improve-bioprocess-workflows-if-applied-correctly.
Rodriguez O, Rosengarten R, Satz A. The Promise of Artificial Intelligence in Healthcare. BioProcess Int. 19(6) 2021; https://lne.e92.mwp.accessdomain.com/manufacturing/information-technology/the-promise-of-ai-in-healthcare.
Corresponding author Oscar Rodriguez is vice president and head of data engineering at Neumora (oscar.rodriguez@neumoratx.com). Adam Roose is vice president of cell and gene commercial strategy at Adjuvant Partners (adam@theaaih.org). Andrea Vuturo is head of communication at AAIH (andrea@vuturo.com). Rafael Rosengarten is CEO at Genialis (rafael@genialis.com). This article was edited from its original form as an informational white paper from the Alliance for Artificial Intelligence in Healthcare.