Bioprocesses traditionally use (fed-)batch cell culture processes for production of recombinant proteins and therapeutics. In batch bioprocessing, material flow is discrete, with a hold step between two unit operations, and product is harvested only once for each unit operation. Batch processes have been studied extensively and optimized through numerous advancements in experimental design (1, 2), monitoring (3–5), measurement techniques (6–9), and control strategies (10–12). However, such processes require large facility footprints for equipment (13) as well as sterilization, load, and harvest steps between individual batches, which in turn decreases their overall space–time yield (STY) (14). Furthermore, many traditional batch processes have shown inconsistent performance in productivity and critical quality attributes (CQAs). Therefore, companies increasingly turn to alternative operation modes for overcoming those process bottlenecks associated with batch culture.
With the introduction of single-use systems, continuous processing offers an attractive alternative to batch processing in terms of facility footprint (for lean manufacturing) and STY. This approach already has been implemented in both the food and chemical industries. However, continuous processing has not been implemented commonly in the biopharmaceutical industry primarily because of strict regulatory interpretations. Both the US Food and Drug Administration (FDA) and the European Medicines Agency (EMA) have defined batches in terms of material quantities rather than mode of operation (15, 16), which could pave the way for accelerated implementation of continuous processing.
The biopharmaceutical industry is beginning to move from batch and fed-batch processing toward continuous processing: bioprocesses in which a continuous flow of material at a steady state is achieved with controlled productivity (17). The main advantages of continuous processing are low production costs, constant product recovery, and increased manufacturing flexibility (18). However, this mode has several disadvantages of its own, from process variability to long development times.
Process variability arises from changes in critical process parameters (CPPs), key performance indicators (KPIs), and critical material attributes (CMAs), all of which in turn affect CQAs. CPPs can change throughout a culture and therefore necessitate different control strategies for reducing process variability. In recent decades, advances in process analytical technology (PAT) and quality by design (QbD) approaches have reduced process development times significantly (19–21). Even though a number of approaches to overcoming process variability do exist, fully continuous processing has yet to be realized in industrial bioprocesses. That is mainly attributable to bottlenecks associated with process variability, infrastructure, and technology transfer.
We postulated the following queries describing what we considered to be the primary obstacles to overcome in an attempt to realize continuous processing in the biopharmaceutical industry:
- Process models are available to identify variability but seldom are used in industrial scenarios. Why are modeling approaches still not transferable to industrial bioprocesses?
- Continuous bioprocessing requires real-time monitoring of process states to ensure robust productivity. Where is the bottleneck to achieving real-time process monitoring?
- Biological variations and time effects should be controlled during continuous processing. How can we effectively control or counteract them?
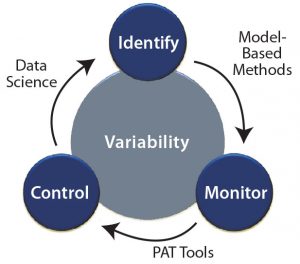
Below, we discuss different modeling approaches, monitoring techniques, and control strategies as enablers to overcoming process variability. Figure 1 is a schematic representation of tackling process variability using different tools.
Overcoming Process Variability
Identification: Process variability associated with CMAs arises from variations in raw material quality, either from batch variations in a supplier’s own manufacturing process or when switching between suppliers. Recently, for example, researchers have combined ultraviolet high-pressure liquid chromatographic (UV-HPLC) data with chemometrics to determine nine main components (CMAs) in the complex matrix of corn steep liquor (CSL) (22).
Variability in KPIs (e.g., cell growth) is of utmost importance for consideration in continuous bioprocess development because of its direct effect on process performance and product quality. But identifying variability in KPIs and controlling it is cumbersome and requires model-based methods. Mechanistic models can be used to describe KPI variability based on process data and mechanistic links (e.g., growth rate) that contain process information describing the state of cells in culture using verifiable data extracted from a bioprocess. Such links usually are based on first-order principles or kinetic equations that describe the general relationships among process parameters (e.g., CPPs) and KPIs.
In recent decades, organizations have made available huge libraries of mechanistic linkages for establishing mechanistic models (23). The main problem with developing such models is that they are nontransferable, which hinders the use of modeling techniques in bioprocess development. A 2017 article described a self-iterative development workflow that identifies and validates mechanistic links for establishing a mechanistic model (23). That workflow includes determining whether mechanistic links from process information can be identified with process data using sensitivity analyses — and if not, it indicates the need for gathering more process data to describe the mechanistic linkages in a process. Such workflows can be used directly at the time of process development for developing a model for bioindustrial scenarios. This workflow has been used to establish a mechanistic model in Chinese hamster ovary (CHO) cell bioprocesses, for which the state of cells and KPIs are determined using an iterative cycle of validation (23).
Process modeling is necessary to identify variations in early stages of process development. Consequently, developers must use automated workflows that generate models in a structured frame, then implement those models to identify sources of process variability. Workflows such as those described above can be integrated throughout an entire product life cycle to generate mechanistic models and easily identify process variability at early stages. Furthermore, modeling workflows are the solution to accelerating technology transfer and vastly reducing development times.
Modeling approaches require real-time data for identifying and monitoring process variations. Real-time monitoring requires timely data collection, transmission, and analysis to capture process variability and predict process performance. Therefore, it is important that a technological and information infrastructure be available for processing data in real time for monitoring process variability.
Monitoring: Measuring, monitoring, modeling, and control (M3C) methodology are the cornerstones of robust bioprocesses.CPPs (e.g., temperature and pH) are measured using standard sensors or soft-sensor software (if a parameter cannot be determined directly). The term soft sensor refers to models that extrapolate process information from indirect measurements (24). For example, a well-established soft sensor for online biomass estimation has been implemented based on mass balances and off-gas analysis (25). Soft sensors for monitoring biomass subpopulations in CHO bioprocesses were reported last year (26).
Modern advanced commercial PAT tools now are available for real-time measurement of CPPs (e.g., substrate concentration) and cell-state variables (e.g., biomass segregation). For example, an online microscope can be used to determine variations in biomass populations (e.g., to distinguish live from dead biomass) in CHO bioprocesses (27). Changes in state variables such as cell lysis during CHO bioprocesses have been analyzed for their impact on cell count, growth kinetics, and productivity (28). In continuous bioprocessing, real-time monitoring of process variability is prerequisite to identifying process deviations and ensuring consistent performance.
Based on modeling techniques and prior knowledge about the state of cells in culture, future predictions of state variables can be made. Observer algorithms (e.g., the Kalman filter) can be used for real-time estimations of state variables and thus for monitoring those variables in real time. Hardware sensors estimate the current state of a variable to monitor it, then the model is used to predict the future states of that variable at set time points. The observer algorithm is used to correct for changes in the state variable. With integrated hardware sensors and models, the entire analysis is facilitated by real-time architecture such as process management systems. Figure 2 shows the observer algorithm for determining the state variable x (e.g., cell concentration) using measurements and model predictions y.
Architecture for performing real-time process monitoring has been developed and implemented in some production bioprocesses. However, to extract the maximum amount of useful information, advanced PAT tools need to be combined with real-time architecture to achieve real-time process monitoring. A recent study combined mechanistic modeling and Raman spectroscopy for real-time monitoring of unmeasurable states in an industrial fed-batch bioprocess (29). Similar approaches have been taken to define the optimal time point of process termination (30, 31) and to monitor the onset of cell lysis (5, 32) for ensuring continuous bioprocessing.
Advanced PAT tools necessarily facilitate data acquisition for modeling approaches, real-time analysis, and feedback of process information, as well as for reduction of error-prone offline measurements. To exploit such tools, real-time architecture for data processing and modeling will be needed. Information from different modeling approaches and PAT tools can be fed back into a process model for real-time monitoring of process variability, thereby enabling continuous processing. Such systems can be integrated directly into early stages of process development, making them highly beneficial for product lifecycle management and technology transfer.

Figure 3: Mechanistic relationship between glucose and lactose uptake rates for tuning between growth and production in an Escherichia coli DE3 bioprocess; dashed line represents the switch between production and growth.
Control: Bioprocesses are dynamic by nature, with constant changes in process conditions and parameters. Biological time effects cause the most variability inside process design spaces. Although that is well known and different approaches have been developed to negate biological time effects, such negation has yet to be realized. Advances in molecular biology pave the way for robust promoter systems that can be used to tune productivity (33, 34) and prevent process variations. Tunable promoter systems have been developed recently and used for removing biological time effects and ensuring robust productivity. Such systems can help maintain cultured cells in a productive state for long durations and render consistent product quality. Furthermore, promoter systems can be used to switch cellular physiology from biomass (growth) to product formation. Figure 3 exemplifies mechanistic relations among substrate uptake rates in a tunable promoter system of Escherichia coli.
The ultimate goal of continuous processing is to keep a production culture healthy over long periods of time and thus lessen or eliminate process variation. Combining tunable promoter systems with model-based approaches can help a system attain constant productivity. Simultaneous control of different feeds for sophisticated strategies has been implemented (35). Furthermore, monitoring specific productivity can be used to determine the optimal time point for harvest to ensure product quality and reduce downstream complications. One mechanistic model has been used for concomitant feed-control strategies to curb process variability (34, 36).
Tunable promoter systems and control strategies can be used to maintain cells in a productive state for the longest possible time and to control consistent product quality, thereby enabling continuous processing. Furthermore, Monte Carlo techniques can be used to simulate process performance within a defined design space and thus enable process engineers to determine the effects of CPP variability on desired product quality. Based on those simulations, control charts can be created to prevent unwanted process deviations and help control process variability within a narrow range.
Our Questions Answered
Biopharmaceutical processes are complex and dynamic by nature, with high degrees of variability that lead to inconsistent and unpredictable process outcomes. To overcome such complexities and understand process variability, bioprocess engineers need a complete knowledge of process parameters and raw material attributes and their effects on quality attributes. We propose the use of modeling workflows, advanced PAT tools, and tunable promoter systems to reduce process variability. To overcome it for continuous processing, we can answer the questions raised above:
Why can’t modeling approaches be transferable to industrial bioprocesses yet? Modeling workflows will be the enabler to generate process models quickly for direct transfer and testing in industrial processes.
Where is the bottleneck to achieving real-time process monitoring? We envision the use of advanced PAT tools — for example, using genetic fingerprinting methods and CQA analysis — and real-time architecture as enablers for real-time process monitoring.
How can we effectively control or counteract biological time effects and variations? Tunable promoter systems are enablers to effectively counteract biological time effects, thereby curbing process variability and providing the essential step toward continuous upstream processing.
Combining all three enablers (modeling, monitoring, and control) to curb process variability will be needed to ensure robust continuous bioprocessing. A cystic fibrosis drug has been produced using a continuous upstream process (37). Such examples highlight the biopharmaceutical industry’s interest in implementation of continuous approaches. Thanks to the available tools and advances, we envision intense incorporation of continuous processing in the biopharmaceutical industry within the coming decades.
References
1 Lim M, et al. Intelligent Bioprocessing for Hemotopoietic Cell Cultures Using Monitoring and Design of Experiments. Biotechnol. Adv. 25(4) 2007: 353–368; doi:10.1016/j.biotechadv.2007.02.002.
2 Toms D, Deardon R, Ungrin M. Climbing the Mountain: Experimental Design for the Efficient Optimization of Stem Cell Bioprocessing. J. Biol. Eng. 11, 2017: 35; doi:10.1186/s13036-017-0078-z.
3 O’Mara P, et al. Staying Alive! Sensors Used for Monitoring Cell Health in Bioreactors. Talanta 176, 2018: 130–139; doi:10.1016/j.talanta.2017.07.088.
4 Claßen J, et al. Spectroscopic Sensors for In-Line Bioprocess Monitoring in Research and Pharmaceutical Industrial Application. Anal. Bioanal. Chem. 409(3) 2017: 651–666; doi:10.1007/s00216-016-0068-x.
5 Rajamanickam V, et al. A Novel Toolbox for E. coli Lysis Monitoring. Anal. Bioanal. Chem. 409(3) 2016: 1–5; doi:10.1007/s00216-016-9907-z.
6 Constantinou A, Polizzi KM. Opportunities for Bioprocess Monitoring Using FRET Biosensors. Biochem. Soc. Trans. 41(5) 2013: 1146–1151; doi:10.1042/BST20130103.
7 Demuth C, et al. Novel Probes for pH and Dissolved Oxygen Measurements in Cultivations from Millilitre to Benchtop Scale. Appl. Microbiol. Biotechnol. 100(9) 2016: 3853–3863; doi:10.1007/s00253-016-7412-0.
8 Faassen SM, Hitzmann B. Fluorescence Spectroscopy and Chemometric Modeling for Bioprocess Monitoring. Sensors (Switzerland) 15(5) 2015: 10271–10291; doi:10.3390/s150510271.
9 Gonzalez-Cabaleiro R, et al. Heterogeneity in Pure Microbial Systems: Experimental Measurements and Modeling. Front. Microbiol. 8, 2017: 1813; doi:10.3389/fmicb.2017.01813.
10 Stanke M, Hitzmann B. Automatic Control of Bioprocesses. Adv. Biochem. Eng. Biotechnol. 132, 2013: 35–63; doi:10.1007/10_2012_167.
11 Ehgartner D, et al. Controlling the Specific Growth Rate Via Biomass Trend Regulation in Filamentous Fungi Bioprocesses. Chem. Eng. Sci. 172, 2017: 32–41; doi:10.1016/j.ces.2017.06.020.
12 Cos O, et al. Operational Strategies, Monitoring, and Control of Heterologous Protein Production in the Methylotrophic Yeast Pichia pastoris Under Different Promoters: A Review. Microb. Cell Fact. 5, 2006: 17; doi:10.1186/1475-2859-5-17.
13 Hernandez R. Continuous Manufacturing: A Changing Processing Paradigm. BioPharm Int. 28, 2015: 20–27.
14 Croughan MS, Konstantinov KB, Cooney C. The Future of Industrial Bioprocessing: Batch or Continuous? Biotechnol. Bioeng. 112(4) 2015: 648–651; doi:10.1002/bit.25529.
15 Section 210.3: Definitions. Code of Fed. Regul. Title 21, Part 210. US Food and Drug Administration: Rockville, MD, 2017; www. accessdata.fda.gov/scripts/cdrh/cfdocs/cfcfr/CFRSearch.cfm?fr=210.3.
16 EMA/CHMP/BWP/187338/2014. Guideline on Process Validation for the Manufacture of Biotechnology-Derived Active Substances and Data to be Provided in the Regulatory Submission. European Medicines Agency: London, UK, 29 April 2016.
17 Rathore AS, et al. Continuous Processing for Production of Biopharmaceuticals. Prep. Biochem. Biotechnol. 45(8) 2015: 836–849; doi:10.1080/10826068.2014.985834.
18 Jungbauer A. Continuous Downstream Processing of Biopharmaceuticals. Trends Biotechnol. 31(8) 2013: 479–492; doi:10.1016/j.tibtech.2013.05.011.
19 Rathore AS, et al. Case Study and Application of Process Analytical Technology (PAT) Towards Bioprocessing: Use of On-Line High-Performance Liquid Chromatography (HPLC) for Making Real-Time Pooling Decisions for Process Chromatography. Biotechnol. Bioeng. 100(2) 2008: 306–316; doi:10.1002/bit.21759.
20 Li M, et al. In Situ Infrared Spectroscopy As a PAT Tool of Great Promise for Real-Time Monitoring of Animal Cell Culture Processes. Austin J. Analyt. Pharm. Chem. 3(2) 2016: 1065.
21 del Val IJ, Kontoravdi C, Nagy JM. Towards the Implementation of Quality By Design to the Production of Therapeutic Monoclonal Antibodies with Desired Glycosylation Patterns. Biotechnol. Prog. 26(6) 2010: 1505–1527; doi:10.1002/btpr.470.
22 Hofer A, Herwig C. Quantitative Determination of Nine Water-Soluble Vitamins in the Complex Matrix of Corn Steep Liquor for Raw Material Quality Assessment. J. Chem. Technol. Biotechnol. 92(8) 2017: 2106–2113; doi:10.1002/jctb.5211.
23 Kroll P, et al. Workflow to Set Up Substantial Target-Oriented Mechanistic Process Models in Bioprocess Engineering. Process Biochem. 2017: 1–13; doi:10.1016/j.procbio.2017.07.017.
24 Luttmann R, et al. Soft Sensors in Bioprocessing: A Status Report and Recommendations. Biotechnol. J. 7(8) 2012: 1040–1048.
25 Wechselberger P, Sagmeister P, Herwig C. Real-Time Estimation of Biomass and Specific Growth Rate in Physiologically Variable Recombinant Fed-Batch Processes. Bioprocess Biosyst. Eng. 36(9) 2013: 1205–1218; doi:10.1007/s00449-012-0848-4.
26 Kroll P, Stelzer IV, Herwig C. Soft Sensor for Monitoring Biomass Subpopulations in Mammalian Cell Culture Processes. Biotechnol. Lett. 1–7, 2017; doi:10.1007/s10529-017-2408-0.
27 Barbau J, Henneghien J. Automated Suspension Cell Culture Monitoring in Stirred Tank Reactors. OVIZIO Imaging Systems SA/NV: Brussels, Belgium, 2017; www.ovizio. com/_dbfiles/lacentrale_files/200/228/Application-Note—iLine-F–Bioconnect-V11dd01102014.pdf.
28 Kroll P, et al. Impact of Cell Lysis on the Description of Cell Growth and Death in Cell Culture. Eng. Life Sci. 17(4) 2017: 440–447; doi:10.1002/elsc.201600088.
29 Golabgir A, Herwig C. Combining Mechanistic Modeling and Raman Spectroscopy for Real-Time Monitoring of Fed-Batch Penicillin Production. Chemie-Ingenieur-Technik 88(6) 2016: 764–776; doi:10.1002/cite.201500101.
30 Eggenreich B, et al. A Combination of HPLC and Automated Data Analysis for Monitoring the Efficiency of High-Pressure Homogenization. Microb. Cell Fact. 16, 2017: 134; doi:10.1186/s12934-017-0749-y.
31 Rajamanickam V, et al. An Automated Data-Driven DSP Development Approach for Glycoproteins from Yeast. Electrophoresis 38(22–23) 2017: 2886–2891; doi:10.1002/elps.201700229.
32 Klein T, et al. Quantification of Cell Lysis During CHO Bioprocesses: Impact on Cell Count, Growth Kinetics, and Productivity. J. Biotechnol. 207, 2015: 67–76.
33 Wurm DJ, Herwig C, Spadiut O. How to Determine Interdependencies of Glucose and Lactose Uptake Rates for Heterologous Protein Production with E. coli. Meth. Mol. Biol. 1586, 2017: 397–408; doi:10.1007/978-1-4939-6887-9_26.
34 Wurm DJ, et al. The E. coli pET Expression System Revisited: Mechanistic Correlation Between Glucose and Lactose Uptake. Appl. Microbiol. Biotechnol. 100(20) 2016: 8721–8729; doi:10.1007/s00253-016-7620-7.
35 Sagmeister P, et al. Tunable Recombinant Protein Expression with E. coli in a Mixed-Feed Environment. Appl. Microbiol. Biotechnol. 98(7) 2014: 2937–2945; doi:10.1007/s00253-013-5445-1.
36 Wurm DJ, et al. Mechanistic Platform Knowledge of Concomitant Sugar Uptake in Escherichia coli BL21(DE3) Strains. Sci. Rep. 7, 2017: 45072; doi:10.1038/srep45072.
37 Estes K, Langer E. Update on Continuous Bioprocessing: From the Industry’s Perception to Reality. Pharm. Technol. 41, 2017: 70–72.
Vignesh Rajamanickam is a project assistant in both the biochemical engineering research division and the Christian Doppler Laboratory for Mechanistic and Physiological Methods for Improved Bioprocesses (Institute of Chemical, Environmental, and Biological Engineering) at the Vienna University of Technology (TU Wien), Gumpendorfer Straße 1a/BH, Fourth Floor, Vienna, Austria; 43-1-58801-166496; vignesh. rajamanickam@tuwien.ac.at. Oliver Spadiut is an assistant professor in TU Wien’s biochemical engineering research division. Corresponding author Christoph Herwig is founder and scientific advisor of Exputec and a bioprocess engineering professor with both the biochemical engineering research division and the Christian Doppler Laboratory for Mechanistic and Physiological Methods for Improved Bioprocesses (Institute of Chemical, Environmental and Biological Engineering) at TU Wien; 43-58801-166400; christoph.herwig@tuwien.ac.at.