In multistep schemes, hydrophobic charge-induction chromatography (HCIC) has been shown to contribute effectively to clearance of Chinese hamster ovary (CHO) host-cell proteins (CHOPs), DNA, and viruses. When used for capture chromatography, HCIC can provide better aggregate clearance than protein A sorbents can. Chen et al. enhanced clearance of aggregates, CHOPs, and product- related impurities by controlling HCIC based on both pH and the presence of binding-promoting salt in the wash and elution buffers used (1). Taken together with our findings presented herein, published results suggest that HCIC could play a broader role in monoclonal antibody (MAb) platform development.
Part 1 of this report (2) provided background and described materials and methods used in our study of MEP HyperCel HCIC sorbent. Here, we conclude by discussing the results of 13 experiments, numbered according to their listings in Table 1 (see Part 1 ) (2). We investigated HCIC control methods as well as the influence of both residence time and feedstock concentration. In addition, we compared isolation results from murine IgG1 from albumin- supplemented and albumin-free growth media. We also purified antibodies representing additional isotypes and species sources. Table 1 summarizes the chromatographic conditions, antibodies, and feedstock compositions for all experiments — with a summary statement of objectives for each experiment or group of experiments (2).
Results and Discussion
Optimizing Mobile-Phase pH and Salt Concentration During Purification of a Monoclonal Murine IgG1: With Experiments 1–4, we hoped to determine optimum pH of elution. We also wanted to determine whether addition of sodium chloride (NaCl) to wash or elution buffers could enhance the resolution of separation, particularly of the target antibody from albumin present in protein-supplemented growth media. We also investigated the influence of NaCl added to the feedstock.
Ultimately, we determined that no addition of binding-promoting salt was required during loading. In fact, other studies using a murine IgG1 (3) have demonstrated that a binding capacity of 37 mg/mL could be achieved when loading with a dilute buffer (25 mM Tris HCl, pH 8.0). Significantly, that binding capacity value was similar for a murine IgG2a (34 mg/mL) loaded under similar conditions. By contrast, efficient binding of murine IgG1 on protein A sorbents typically requires adding a binding-promoting salt and loading at pH ≥8.5 (4). No such enhanced binding conditions are required for binding murine IgG1 on MEP HyperCel sorbent. That illustrates the broad selectivity of HCIC compared with protein A chromatography. Additional examples are considered below.
Because of the low IgG titer of our clarified cell-culture supernatant, for Experiments 1–4 we used feedstock concentrated 30-fold with tangential- flow filtration (TFF) using a 50-kDa membrane (see Materials and Methods, 2). For Experiments 5–8, as described below, we used clarified supernatant without concentration to assess the feasibility of working directly with dilute feedstock. As noted in Table 1 (2), Experiment 8 was conducted at increased scale.
Evaluation of Preelution Wash Steps (Experiment 1): In this initial experiment (Figure 2), we used a limited number of pH elution steps; subsequent experiments involved a more comprehensive pH step-elution sequence. Experiment 1 also included preelution wash steps with pure water (W2) and with 25 mM sodium caprylate in equilibration buffer (W3). Wash steps W2 and W3 followed the initial postload wash with equilibration buffer (W1).
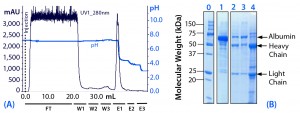
Figure 2: (a) Purification chromatogram of monoclonal mouse IgG1 from cell culture supernatant using MEP HyperCel sorbent with brief pH-step-elution sequence (Experiment 1)*; (b) sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) in reducing conditions**Figure 2 footnotes * 0.46-cm ID Ă— 5-cm bed height column with 0.83 mL resin and phosphate-buffered saline (PBS) equilibration buffer, pH 7.4; 20 mL sample (concentrated cell culture supernatant containing mouse IgG1 at 1.5 mg/mL) injected at a linear flow rate of 60 cm/h (residence time 5 minutes); column washed with PBS at pH 7.4 (W1), deionized water (W2) and 25 mM sodium caprylate (W3); antibodies eluted with a 50 mM sodium acetate buffers sequence, pH 4.5 (E1), pH 4.0 (E2), and pH 3.0 (E3) ** Lane 0 = molecular weight (MW) standards; lane 1 = cell culture supernatant; lane 2 = deionized water wash fraction (W2); lane 3 = sodium caprylate wash fraction (W3); lane 4 = main elution pool eluted at pH 4.5 (E1); HC = heavy chain, LC= light chain
For applications in which the feedstock contains albumin, those additional wash steps can be useful for selectively desorbing it and other impurities that are less strongly retained than the target immunoglobulin. Desorption is prompted by the reduced conductivity of pure water. Sodium caprylate then provides mild, detergent-like effects to prompt desorption while also operating as a complexing agent for albumin (5).
That said, those supplementary wash steps can prompt elution of some target antibody, reducing yield. We judged this to be the case in Experiment 1 (Figure 2, lanes 2 and 3). Thus, we did not use washes W2 and W3 in subsequent experiments, instead pursuing other strategies to resolve antibody from albumin.
Eluted at pH 4.5, the principal target fraction (E1) was judged to be <90% pure. We observed residual albumin and transferrin. Thus, in subsequent experiments, the elution sequence included additional steps at higher pH levels to provide less aggressive elution conditions and allow more nuanced control of the separation.
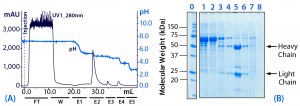
Figure 3: (a) Purification chromatogram of monoclonal mouse IgG1 from cell culture supernatant using MEP HyperCel sorbent (Experiment 2)*; (b) sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) in reducing conditions**Figure 3 footnotes
* 0.46-cm ID Ă— 5-cm bed height column with 0.83 mL resin) and phosphate-buffered saline (PBS) equilibration buffer, pH 7.4; 10 mL sample (concentrated cell culture supernatant containing mouse IgG1 at 1.5 mg/mL) injected at a linear flow rate of 40 cm/h (residence time 7.5 minutes); column washed with PBS, pH 7.4 (W); antibodies eluted with 100 mM sodium acetate buffers, pH 5.5 (E1), pH 5.2 (E2), 50 mM sodium acetate buffers, pH 4.8 (E3), pH 4.0 (E4), and pH 3.0 (E5)
** Lane 0 = molecular weight (MW) standards; lane 1 = cell culture supernatant; lane 2 = flow through (FT); lane 3 = wash (W); lanes 4–8 = elution pools E1–E5, respectively; HC = heavy chain, LC = light chain
Optimization of Elution pH (Experiment 2): As Figure 3 shows, the pH step-elution sequence ranged from pH 5.5 to pH 3.0, with steps of 0.3 or 0.4 units between pH 5.5 and pH 4.8. The principal antibody fraction eluted at pH 5.2 (E2). As anticipated, preferential elution of albumin was favored at higher pH (less aggressive elution conditions). Nevertheless, we found a significant quantity of target antibody in fractions collected at pH 5.5 (E1) and 4.8 (E3), where it coeluted with albumin, transferrin, and other impurities. A range of strongly bound impurities eluted at pH 4.0 (E4).
Based on this experiment, we concluded that HCIC optimization based solely on pH would be insufficient to provide a product of 90% purity at 90% yield. Thus, we conducted additional optimization based on salt concentration in the mobile phase. Specifically, we added to the mobile phase a binding-promoting salt to prompt selective retention of the antibody under conditions at which albumin and transferrin (less hydrophobic proteins) would be unretained.
Optimization of NaCl Concentration for Binding and Elution (Experiments 3 and 4): To develop an overview of how binding-promoting salt influences all components of the feedstock, we added 1M NaCl to all buffers and to the feedstock in Experiment 3. Here, we combined the feedstock used in Experiments 1 and 2 with an equal volume of equilibration buffer containing 2M NaCl for a final concentration of 1M NaCl and 0.75Â mg IgG per milliliter of feed.
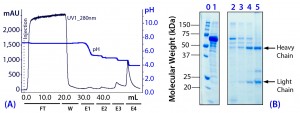
Figure 4: (a) Purification chromatogram of monoclonal IgG1 from cell culture supernatant using MEP HyperCel sorbent (Experiment 3)*; (b) sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) in reducing conditions**Figure 4 footnotes
* 0.46-cm ID Ă— 5-cm bed height column with 0.83-mL resin and phosphate-buffered saline (PBS) equilibration buffer, pH 7.4, NaCl 1M; 20 mL sample (concentrated cell culture supernatant containing mouse IgG1 at 1.5 mg/mL diluted 1/2 (v/v) in PBS, pH 7.4, NaCl 2M) injected at a linear flow rate of 40 cm/h (residence time 7.5 minutes); column washed with PBS at pH 7.4, NaCl 1M (W); antibodies eluted with 100 mM sodium acetate buffers containing 1M NaCl, pH 5.5 (E1), pH 5.2 (E2), 50 mM sodium acetate buffers containing 1M NaCl pH 4.8 (E3), and pH 4.0 (E4)
** Lane 0 = molecular weight (MW) standards; lane 1 = cell culture supernatant; lanes 2–5 = elution pools eluted with buffers containing 1M NaCl at pH 5.5 (E1), pH 5.2 (E2), pH 4.8 (E3), and pH4.0 (E4) respectively; HC = heavy chain, LC = light chain
As Figure 4 shows, this strategy increased retention of all components. However, we determined that adding NaCl had a more pronounced effect on antibody retention than on retention of albumin and transferrin. For example, the principal antibody fraction now emerged at pH 4.8 (E3) and pH 4.0 (E4). By contrast, the principal antibody fraction had emerged at pH 5.2 in Experiment 2 (see Figure 3). Significant elution of albumin and transferrin continued to occur at pH 5.5 even with added NaCl present.
Moreover, a critical observation in Experiment 3 was that only a trace quantity of antibody eluted at pH 5.5 in the presence of NaCl. These findings suggested that selective desorption of albumin and/or transferrin could be prompted at pH 5.5 in the presence of NaCl, while target antibody would be selectively retained. We designed Experiment 4 to further evaluate and exploit these observations.
During Experiment 4, we altered several variables relative to the conditions in Experiment 3. First, we did not augment the feedstock or equilibration buffer with NaCl. Next, we added supplementary NaCl only to the pH 5.5 buffer (E1). Our objective here was to prompt antibody retention at that pH and salt concentration while allowing selective elution of albumin and/or transferrin. Moreover, we reduced the NaCl concentration in the pH 5.5 buffer to 0.5 M (E1) to determine whether that concentration would be sufficient to prompt selective antibody retention. Finally, we designed this experiment to determine whether efficient antibody elution could be achieved with pH reduced by an additional increment (to pH 5.0) using buffer without added NaCl (E2).
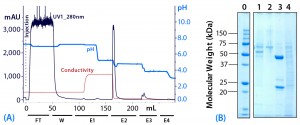
Figure 5: (a) Purification chromatogram of monoclonal IgG1 from cell culture supernatant using MEP HyperCel sorbent in an optimized elution protocol (Experiment 4)*; (b) sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) in reducing conditions**Figure 5 footnotes
* 1.0-cm ID Ă— 9.7-cm bed height column with 7.6 mL resin and phosphate-buffered saline (PBS) equilibration buffer, pH 7.4; 45 mL sample (concentrated cell culture supernatant containing mouse IgG1 at 1.5 mg/mL); sample volume injected at a linear flow rate of 80 cm/h (residence time 7.5 minutes); column washed with PBS at pH 7.4; elution step E1 = 100 mM sodium acetate buffer containing 0.5 M NaCl, pH 5.5, E2 = 50 mM sodium acetate buffer pH 5.0, E3 = pH 4.0, and E4 = pH 3.0
** Lane 0 = molecular weight (MW) standards; lane 1 = wash fraction; lanes 2–4 = elution pools (E1, E2, and E3, respectively)
As Figure 5 shows, the fraction eluted at pH 5.5 using buffer augmented with 0.5 M NaCl (E1) contained little or no target antibody. Albumin and transferrin appeared principally in the postload wash with equilibration buffer (W). Remaining early eluting impurities appeared, as intended, in the fraction eluted at pH 5.5 with buffer containing 0.5 M NaCl (E1). The target antibody was recovered at pH 5.0 in NaCl-free buffer (E2).
Based on UV absorbance (A280) of the target fraction, the antibody concentration in the target fraction from Experiment 4 was significantly greater than in elution fractions from Experiment 3. This is particularly noteworthy because column loading was greater in Experiment 3 than in Experiment 4 (Table 1) (2). Based on analysis with sodium-dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), only trace quantities of albumin or transferrin appeared in this fraction (Figure 5). Based on size-exclusion high-performance liquid chromatography (HPLC) analysis, we judged that fraction to be 96% pure. Recovery of applied antibody in this fraction amounted to 93%. We conducted further elution steps at pHÂ 4.0 and pH 3.0, but the resulting fractions contained no significant quantities of target antibody. However, we observed elution of a mixture of impurity components at pH 4.0 (E3).
Although this work focused on resolution of antibody from less- strongly retained impurities, HCIC control based on both pH and the presence of a binding-promoting salt also has been used to facilitate resolution of monomeric antibody from more strongly retained impurities: antibody aggregates (1). Other investigators have studied the influence of both pH and binding-promoting salt on HCIC for chromatography of both antibody and nonantibody proteins (6).
For Experiment 4, we chose a residence time of 7.5 minutes (Figure 5) (Table 1) (2). Related experiments (data not shown) revealed that with residence time decreased to 3.8Â minutes and all other variables unchanged, product of similar purity (97%) was recovered but at a significantly lower recovery rate of ~78%. For purposes of approximation, we assumed a linear relationship between residence time and recovery. Based on that approximation, we estimated that the target recovery value (90%) could be achieved with a residence time of 6.8 minutes using the current feedstock.
Influence of recovery on residence time will be application and feedstock dependent. We conducted further studies of residence time’s influence using unconcentrated, dilute feedstock. These experiments are identified in Tables 1 (2) and 2. As discussed below, loading of dilute feedstock permitted reduced residence times while maintaining target recovery values (≥90%).
Influence of Residence Time (Experiments 5–8): In the next four experiments, we explored the influence of residence time and feedstock concentration on chromatographic performance during purification of a monoclonal murine IgG1.
Influence of Residence Time Using Low-Concentration Feedstock (Experiments 5–7): We applied the optimized buffer formulations identified  during Experiment 4, but using unconcentrated feedstock (~0.043 mg murine IgG1 per milliliter of feed) and sample loading reduced residence times (2.1, 3.0, and 5.0 minutes, compared with 7.5 minutes in Experiment 4). As Tables 1 (2) and 2 show, increased volumes of feedstock correspond to loads ranging from 4.2 to 10.1 mg IgG per milliliter of sorbent (compared with 8.9 mg of IgG/mL during Experiment 4).

Table 2: Influence of residence time on chromatographic performance during chromatography with MEP HyperCel sorbent — comparing unconcentrated and concentrated feedstocks in Experiments 4–8; all feedstocks came from cultures using hybridoma serum-free media (HSFM) supplemented with transferrin (10 µg/mL), insulin (10 µg/mL), and Albumax lipid-rich bovine serum albumin (concentration proprietary to Abbott).
As Table 2 shows, residence time could be reduced to as low as three minutes while still achieving ≥90% recovery and ≥90% purity. Such behavior is consistent with the reduced diffusion limitations that prevail when feedstock is less concentrated. The high purity values achieved are particularly significant because the feedstock was not subjected to preliminary TFF. So this feedstock would be expected to contain low–molecular-weight impurities in greater abundance. Given these findings, we next repeated the experiment at increased scale.

Figure 6: (a) Purification chromatogram of monoclonal IgG1 from an unconcentrated cell culture supernatant using MEP HyperCel sorbent (Experiment 8*); (b) sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) in reducing conditions (pH 5.0)**Figure 6 footnotes
* 5.0-cm ID × 17.5-cm bed height column with 343 mL resin and phosphate-buffered saline (PBS) equilibration buffer, pH 7.4; 28 L sample (cell culture supernatant containing mouse IgG1 at 43 µg/mL) injected at a linear flow rate of 230 cm/h (residence time, 4.6 minutes) and eluted at 300 cm/h (residence time 3.5 minutes); column washed with PBS at pH 7.4; elution step E1 = 100 mM sodium acetate buffer containing 0.5 M NaCl at pH 5.5, E2 = 50 mM sodium acetate buffer at pH 5.0, E3 at pH 4.0, and E4 at pH 3.0
** Lane 0 = molecular weight (MW) standards; lane 1 = load; lane 2 = elution pool; HC = heavy chain, LC = light chain
Purification of a Monoclonal Murine IgG1 from Dilute Feedstock at Increased Scale (Experiment 8): As shown in Tables 1 (2) and 3 and Figure 6, we conducted the next experiment with a residence time of 4.6 minutes during loading and 3.5 minutes during elution. We applied 28 L of unconcentrated murine IgG1 feedstock (0.041 mg/mL) to a 343-mL column of MEP HyperCel sorbent (equivalent to 3.4 mg IgG/mL sorbent). Our chromatographic sequence and strategy were as applied during Experiments 4–7 (Tables 1 and 2) (2).
As Table 3 shows, we judged the purity of the target fraction (E2, eluted at pH 5.0) to be 97% based on size- exclusion HPLC, with 97% recovery. Those values are of further interest in connection with the general features of the chromatogram in Figure 6. In particular, notice that the preceding step (E1, pH 5.5 containing 0.5M NaCl) prompts elution of a significant quantity of UV-absorbing material. Likewise, the step following elution of the target fraction (E3, pH 4.0), also prompts elution of a significant quantity of UV-absorbing material. Given the high recovery in the target fraction, this process clearly provided good selectivity with little antibody loss in steps proceeding and following recovery of the target fraction. Table 3 presents additional data concerning the feedstock, target fraction, and purification achieved.

Table 3: Purification achieved during Experiment 8 with chromatography of 28 L of unconcentrated feedstock on MEP HyperCel sorbent
Although high expression levels have become the norm in fully developed processes, low expression levels are often encountered during research and developmental phases of studies with new antibodies. The flexibility for efficient use of HCIC with unconcentrated, dilute feedstocks can be helpful in such settings. It is also useful to revisit the notion of diverse “platform” approaches to antibody purification. They may have utility in therapeutic, diagnostic, or research applications at various stages of development. Finally, findings concerning the influence of residence time on chromatographic performance should be of general value because minimizing process time is important to both laboratories and production suites.

Table 4: Purification achieved following chromatography of different feedstocks using MEP HyperCel sorbent and the optimized protocol applied in Experiment 4; all murine experiments but 9 used the same clone (murine IgG1 (1)). Experiment 9 used another clone (murine IgG1 (2)); culture medium A = hybridoma serum-free media (HSFM) supplemented with transferrin (10 µg/mL), insulin (10 µg/mL), and Albumax lipid-rich BSA (concentration proprietary to Abbott); culture medium B = HSFM supplemented with Pluoronic F-68 surfactant (0.05%); culture medium C = Iscove’s modified Dulbecco’s medium (IMDM) supplemented with fetal bovine serum (10%)
Purification of Additional Murine MAbs and an Ovine Monoclonal IgG1 Using Feedstocks Derived from Different Growth Media Formulations (Experiments 9–12): We conducted chromatography of additional MAbs to further explore the broad selectivity and general applicability of HCIC on MEP HyperCel sorbent. As Table 4 shows, we were particularly interested in observing its ability to achieve binding without needing high pH or addition of a binding-promoting salt with species sources or isotypes that are poorly retained on protein A. Accordingly, we worked with an additional murine IgG1 antibody. For comparison, we included a murine IgG2a, an isotype that is well retained on protein A sorbents (3). We included ovine IgG1 because it is also poorly retained on protein A sorbents (7). And finally, we tested different growth media formulations.
For these experiments, we again used the optimized wash-and-elution sequence. As Table 4 shows, we concentrated the feedstocks 30-fold using TFF before chromatography, using different IgG concentration and/or residence times for purifying the different isotypes. Our findings here are consistent with those discussed above and indicate that residence time and feedstock concentration can influence IgG recovery (Table 4). Significantly, we recovered 93–98% pure product, determined by size-exclusion HPLC analysis. As discussed below, we anticipate that enhanced recovery could be achieved with appropriate adjustment of residence time and/or feedstock concentration.
As Table 4 shows, residence time during Experiment 9 equaled that in Experiment 4, with 1.7-fold greater IgG concentration (2.6 mg/mL) in the feedstock. Recovery (89%) was incrementally below the value achieved in Experiment 4 and also incrementally below the target value (≥90%). These findings are consistent with those discussed above indicating dependence of recovery on feedstock concentration. A modest change in residence time or feedstock concentration should provide required recovery.
For a murine IgG2a (Experiment 10), we used a residence time 67% of that in Experiment 4 and achieved only 70% recovery. When we made a similar reduction in residence time (70%) between Experiment 6 and 7 (Table 2), recovery declined from 96% to <80%. Repeating Experiment 10 with an appropriately increased residence time probably would give enhanced recovery in the target range.
In Experiment 11, we evaluated the effect of cell growth medium on purity and IgG recovery using the same murine IgG1 MAb as in Experiments 1–8. This experiment used feedstock derived from serum-free/albumin-free growth medium containing transferrin and insulin at 10 µg/mL along with Pluronic F-68 surfactant (at 0.05% v/v) as an antishear agent to compensate for the absence of albumin. Despite the relatively high IgG concentration (2.8 mg/mL) and short residence time (4.3 minutes), recovery was 100% and product purity was 98%.
That finding is reasonable given the substantially lower concentration of non- IgG protein present in the feedstock. A lower total protein concentration would be expected to favor reduced pore diffusion constraints for the target IgG. Likewise, the absence of albumin would favor reduced binding competition for the chromatographic ligand. As a practical matter, the high purity and recovery values achieved should be of particular interest to readers working with protein-free or chemically defined growth media for therapeutic production.
We evaluated the ability to capture and recover an antibody species other than murine using an ovine IgG1 (Experiment 12) derived from growth medium supplemented with fetal bovine serum. Significantly, we isolated product of 98% purity from this complex feedstock with a relatively short residence time (3Â minutes). Such a combination of short residence time and abundant nonantibody protein in FBS- supplemented growth medium would be expected to limit recovery (here only 65%). As with previous experiments with low recoveries, repetition with appropriately increased residence time should improve that result.
Overall, strategies for bringing recovery values into the target range (>90%) appear straightforward. Dependence of recovery on both residence time and feedstock concentration is particularly clear in the data from Experiments 4–12. Findings for isolation of murine IgG1 from albumin-free growth medium (Experiment 11) are particularly encouraging in connection with efficient purification of high-purity product from feedstocks involving protein-free or chemically defined growth media. Even for more complex, challenging feedstocks, it should be possible to achieve ≥90% high-purity product recovery using a residence time ≤8 minutes.
Proof of Concept
We used HCIC on MEP HyperCel sorbent for capture and purification of monoclonal murine IgG1, an isotype and species-source known to be poorly retained on protein A sorbents. We studied isolation from both concentrated and unconcentrated feedstocks, most derived from serum-free growth media supplemented with albumin, transferrin, and insulin. We also investigated the influence of residence time and feedstock concentration on chromatographic performance. And we optimized HCIC based on both elution pH and the presence of a binding- promoting salt in preelution wash and elution buffers.
With an optimized chromatographic sequence, isolation of target antibody from protein-supplemented growth media yielded product of 93–98% purity in seven experiments, all based on a single chromatographic step. Among those seven experiments, four used unconcentrated feedstock and three used concentrated feedstock. In the former case, recovery values ranged 95–97% for all experiments using residence times ≥3 minutes during loading. With concentrated feedstocks, recovery values ranged 89–93% with a residence time of 7.5 minutes during loading. We believe that a modest increase in residence time would have raised recovery to 90% (the defined objective) for the experiment in which it fell slightly short of that value (89%). The ability to shorten residence times with unconcentrated feedstock is consistent with the reduced diffusion limitations that prevail when feedstock concentration is reduced.
In a parallel experiment conducted to isolate murine IgG1 from concentrated feedstock derived from serum-free, albumin-free growth medium (Experiment 11), we isolated product of 98% purity at 100% recovery using a residence time of only 4.3 minutes. That finding is consistent with the notion that lower total protein concentration in the feedstock would favor reduced pore diffusion constraints for the target IgG. Likewise, the absence of albumin would lessen binding competition for the chromatographic ligand. Findings from work with serum-free, albumin-free growth medium should be of general interest to developers isolating MAbs from protein-free and chemically defined growth media in manufacturing biotherapeutics.
Finally, our studies included isolation of a murine IgG2a and an ovine IgG1 from concentrated feedstocks. We isolated murine IgG2a at 94% purity and ovine IgG1 at 98%purity. The latter is particularly significant because that feedstock came from FBS-supplemented growth medium. However, recovery values fell short of the target value (90%) in both experiments: 70% recovery for murine IgG2a and 65% recovery for ovine IgG1. Based on studies with murine IgG1, the low recovery values achieved probably resulted from insufficient residence time during loading. It is reasonable to anticipate that increased recovery values would be achieved at increased residence times.
Broadly speaking, these findings demonstrate the applicability of HCIC to MAb recovery from feedstocks ranging in complexity from serum- supplemented to serum-free and albumin-free growth media. Although we focused on isolation of MAbs that are poorly retained on protein A sorbents, other studies cited in Part 1 show that HCIC also has been of value in purifying MAbs that are customarily purified using protein A sorbents. Taken together with those reports, our findings demonstrate that HCIC can be useful in development of purification schemes for isolating antibodies in research, diagnostic, and therapeutic applications.
References, Part 2
1 Chen J, et al. The Distinctive Separation Attributes of Mixed-Mode Resins and Their Application in Monoclonal Antibody Downstream Purification Process. J. Chromatogr. A 1217(2) 2010: 216–224.
2 Chaudoreille S, et al. Optimization and Scale-Up of HCIC-Based MAb Purification Processes, Part 1. BioProcess Int. 13(5) 2015: 28–33, 46.
3 MEP HyperCel Sorbent for Hydrophobic Charge Induction Chromatography. Pall Life Sciences: Port Washington, NY, 2004.
4 Gagnon P. Chapter 9: Protein A Affinity Chromatography. Purification Tools for Monoclonal Antibodies. Validated Biosystems: Tucson, AZ, 1996; 162.
5 Johnston A, et al. Low pH, Caprylate Incubation As a Second Viral Inactivation Step in the Manufacture of Albumin: Parametric and Validation Studies. Biologicals 31(3) 2003: 213– 221.
6 Ghose S, Hubbard B, Cramer SM. Evaluation and Comparison of Alternatives to Protein A Chromatography. J. Chromatogr. A 28 July 2006: 144–152.
7 Huse K, Bohme HJ, Scholz GH. Purification of Antibodies By Affinity Chromatography. J. Biochem. Biophys. Meth. 51(3) 2002: 217–231.
At the time this work was done, Stéphanie Chaudoreille was an R&D scientist, and Patrick Santambien was a senior R&D manager at Pall Life Sciences, 48, Avenue des Genottes, 95800 Cergy St. Christophe, France. Natalie Bohrer is a scientist in global protein sciences at Abbvie, 1 North Waukegan Road, North Chicago, IL, 60064. René Gantier is R&D director of biopharm applications, and Warren Schwartz is senior technical director of chromatography at Pall Life Sciences, 20 Walkup Drive, Westborough, MA 01581. Corresponding author Steven P. Allen is manager of biologics process design Abbott Diagnostics Division, 100 Abbott Park Road, Abbott Park, IL 60064; 1-224-668-1006; steven.allen@abbott.com.

