Most biopharmaceutical production platforms are based on fed-batch cell culture protocols, which can support high volumetric productivity while maintaining low operational complexity (1). The industry is interested in developing or refining high-titer cell culture processes to meet increasing market demands and reduce manufacturing costs (2). Although advancements in cell engineering have enabled development of high-performing recombinant cell lines (3,4,5,6), improvements in cell culture media and process parameter settings are required to realize the maximum production potentials of those cells (7,–8).
In a fed-batch process, a basal medium supports initial growth and production, and a feed medium prevents depletion of nutrients and sustains the production phase (9). Those media can contain different assortments of nutrients to accommodate the distinct metabolic requirements during different production phases. Process parameter settings — including feeding strategy and control parameters — define the chemical and physical environments suitable for cell growth and protein production. Optimization of a fed-batch process can be achieved by development efforts addressing one or more of three major elements: basal medium, feed medium, and process settings. Given the large sets of variables in these systems, establishing a cost- and time-efficient approach for process optimization is desired but challenging.
PRODUCT FOCUS: ANTIBODIES AND OTHER PROTEINS
PROCESS FOCUS: PRODUCTION
WHO SHOULD READ: ANALYTICAL AND PROCESS DEVELOPMENT PERSONNEL
KEYWORDS: MEDIA, SUPPLEMENTS, CHO CELLS, DESIGN OF EXPERIMENTS, TITER
LEVEL: INTERMEDIATE
Through a study aiming to upgrade a fed-batch process, we observed that basal and feed media have interrelated impacts on process outcomes (a “pairing effect”). So we required a combined optimization of basal and feed media to fulfill our objective. Here we discuss a rationally integrated approach that best serves the practices of fed-batch cell culture process optimization.
Materials and MethodsChinese Hamster Ovary Cells: We used a recombinant GS-CHO cell line (both the CHOK1SV host cell line and glutamine synthetase expression vector licensed from Lonza Biologics) expressing a monoclonal antibody as the model cell line in our study.
Existing Fed-Batch Cell Culture Process: Development of the existing control fed-batch process has been detailed elsewhere (10). Briefly, a chemically defined medium was chosen as the basal medium, and a chemically defined feed medium was developed through a multiround statistical design of experiment (DoE) study. The process was successfully applied to both NS0 and CHO cell cultures.
Optimization of the Process: We conducted our optimization studies using either shake flasks (Corning) or Cellferm-Pro bioreactors (DASGIP BioTools). Cells were adapted over at least three passages into the test medium variants before evaluation in cell growth performance assays. Initial bioreactor culture volume was 500 mL. We controlled pH at 7.0 ± 0.05 and dissolved oxygen (DO) at 30%. Culture conditions followed the existing control process except for our designed variables. We measured cell densities and viabilities using a Vi-Cell counter (Beckman Coulter) and measured concentrations of glucose, ammonia, and lactate with a BioProfile analyzer (Nova Biomedical). We determined monoclonal antibody titers using a POROS 20-µm protein A ID cartridge affinity column (Life Technologies) controlled by a Waters 2695 Alliance system with 2998 photodiode array detector set at 210 nm (Waters), using a purified IgG standard to establish the standard curves in each measurement.
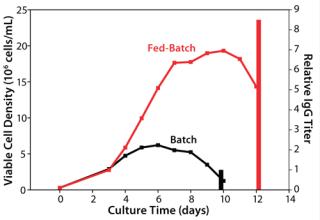
ResultsBaseline Performance of the Control Process: We assessed our existing control process using the model cell line to establish baseline performance. As Figure 1 shows, the fed-batch process reached a peak viable cell (vc) density of 19 × 106 cells/mL, and the batch process (run as a control without feeding) had a peak viable cell density of 7 × 106 cells/mL. The feed in this process demonstrated a high baseline performance by yielding an 8.5-fold titer increase over the batch control process. The IgG titer from our fed-batch process reached the multigram per liter range, which is regarded in the industry as high for a platform process.
Optimization Workflow:Figure 2 depicts our optimization workflow. This project included two major phases: Phase 1 dealt with basal medium development following a three-step DoE path, and phase 2 involved a feed DoE study followed by a combined feed and process optimization.
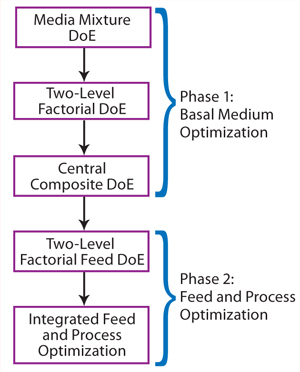
Figure 2:
We conducted basal medium optimization through three steps of DoE experiments. First, we used a media mixture DoE study to identify a basal medium formulation as a starting point for medium optimization. Our second step was a two-level factorial DoE study testing the effects of multiple nutrient groups supplementing the basal medium. The final step was a central composite DoE to determine the optimal concentrations of functional nutrient groups identified in the previous step.
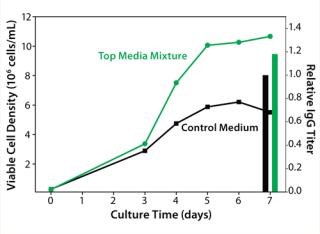
The media mixture layout we designed using Design-Expert software (Stat-Ease), consisted of 15 unique mixtures of four chemically defined prototype formulations. We evaluated our media mixtures with a batch cell growth performance assay. A top media mixture was identified that outperformed the control basal medium by giving 80% higher peak cell densities and 20% higher IgG titers (Figure 3).
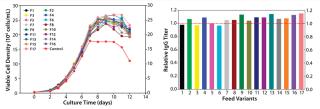
In our two-level factorial DoE study, we selected five nutrient groups to evaluate how they affect protein titer when used as basal medium supplements at two concentrations (added or not) to the top media mixture. We designed a total of 16 basal medium variants with various combinations of those five nutrient groups and assessed them in a fed-batch cell growth performance assay using the existing control feed for all conditions.
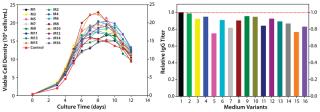
As Figure 4 (LEFT) shows, the cell cultures exhibited a wide range of growth responses to supplementation of the nutrient groups. Multiple supplemented media supported higher peak cell densities than the control condition. However, their boosted cell growth did not translate to a gain in IgG titer; none of the medium variants improved IgG titer over the control (Figure 4, RIGHT). That was contradictory to the previous batch culture evaluation of media mixtures in which improved IgG titers were observed, suggesting that the feed may have been playing a dominant role in IgG production for this process. Furthermore, statistical analyses of the results revealed that two nutrient groups had a negative effect and one nutrient group had a positive effect on IgG titer (p < 0.05).
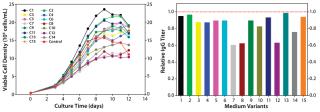
In a central composite DoE study, we regrouped select nutrient groups and tested them at five different concentrations encompassing a wider range than our two-level factorial study. We designed 15 medium variants for evaluation of three nutrient groups. All variants were tested in a fed-batch growth performance assay to which the original control feed was applied. As Figure 5 shows, this round of optimization improved cell growth; however, that did not translate into increased IgG titer.
Cell growth was largely improved through basal medium optimization. By contrast, IgG-specific productivity — which characterizes average productivity per cell — was drastically reduced (data not shown). So an increase in titer was not achieved. Optimally in a fed-batch cell culture process, the basal medium supports cells through their growth phase and thus plays a major role in cell growth, whereas the feed medium sustains cells through the production phase for a primary contribution of increasing IgG productivity. When basal medium optimization improved cell growth, we hypothesized that a corresponding feed optimization would be required to increase the IgG titer of our fed-batch process. Further, we hypothesized that process parameters might need to be altered to realize the full potential of our new medium and feed combination.
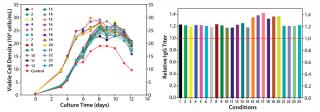
Feed Optimization: In this project, we optimized feed by testing nutrient groups as supplements to the existing feed. We selected five nutrient groups for screening at two levels: high level (with supplements added back to the control feed) and low level (the control feed without supplementation). A total of 17 feed variants reflected different combinations of supplementing the nutrient groups. In the fed-batch cell growth performance assay, we seeded all conditions in the same basal medium (the top medium identified in phase 1), feeding each one with the designated feed variant.
As Figure 6 shows, multiple test conditions outperformed the control fed-batch process with regard to IgG titer, with the top condition demonstrating a 15% increase. This improvement suggested that the fed-batch process may benefit from the pairing effect described above. The control feed was originally developed to specifically match the original basal medium, which gave a high baseline performance (11). After basal medium optimization, feed modification became necessary to better match the new basal medium for superior process outcome. We conducted statistical analyses and identified two nutrient groups with significant positive effects on IgG production (p < 0.05).
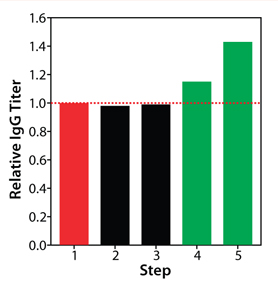
Our final study was an integrated feed and process optimization. We designed five feed variants to test different concentrations of nutrient groups identified as having a significant positive effect on protein titer in our previous experiment. Using a fed-batch growth performance assay, we tested these variants in conjunction with multiple process parameters: seeding density, feeding schedule, and temperature shift. We seeded all conditions in the same basal medium, feeding each condition with the designated variant under different process settings.
As Figure 7 depicts, this round of integrated optimization further improved IgG titer, with the top condition 40% higher than the control fed-batch process. This titer improvement could be primarily linked to increased IgG-specific productivity (data not shown); the accompanying cell growth augmentation was only marginal. This study involved an integrated optimization combining both feed and process optimization in one step. Although feed development and process optimization are usually addressed in separate studies, this integrated approach can be used to deliver superior results in a shortened period.