Mathematical modeling has been widely used in microbiology and biotechnology for several decades. The main objective of modeling is to find optimal conditions for microbial growth and biosynthesis of useful metabolites.
We modified the well-known equation of Perth–Marr (1) — proposed to calculate the energy consumption of a substrate— to analyze the energy consumption by cells for growth and viability maintenance. Our study includes that theory along with our own development. Our initial modeling work was carried out with Yersinia, Pseudomonas, Pasteurella, and Salmonella. For those studies, we created structured, unstructured, and general models (2, 3).
Briefly, we now propose a general model that can be described as follows. Figure 1 shows a simplified scheme of the cell cycle in prokaryotes. Positions 1 and 5 (6) correspond to the boundary points of the cell cycle — a cell of zero age. In the literature, such cells are called resting or dormant, and we call them stable cells, Xst. For bacteria, that corresponds to growth phase B, and for eukaryotic cells it is the phase of G0 and G1. Any intermediate position (2, 3, 4) and an infinite number of positions between them corresponds to dividing cells Xdiv. Such cells are most sensitive to physical, chemical, mechanical, and other influences.
PRODUCT FOCUS: BACTERIA-EXPRESSED PROTEINS, METABOLITES
PROCESS FOCUS: PRODUCTION, FERMENTATION
WHO SHOULD READ: PROCESS DEVELOPMENT AND MANUFACTURING
KEYWORDS: BIOMASS, CELL VIABILITY, SUBSTRATES, MATHEMATICAL MODELING
LEVEL: ADVANCED
Equations 1–2
Equation 1
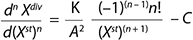
Equation 2
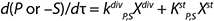
For substrates and metabolites, we suggest
Definitions
LGP: logarithmic growth phase
GIP: growth inhibition phase
S: substrate
X: biomass
τ: time
P: products
dP/dτ: absolute rate of product synthesis;
Q = – dS/dτ: absolute rate of substrate consumption
QO2: oxygen mass exchange rate, mmole O2/(volume units per time units)
J: stochiometric factor of energy substrate (S) oxydation, Joule S/mmole O2
µ: specific growth rate of biomass X per (hour or days)
q = Q/X or q = (1/X) × dP/dτ: a specific rate of substrate use or product synthesis
a: a trophic coefficient, amount of energy substrate consumed for the synthesis of a biomass unit
f: amount of energy substrate accumulated in biomass X during cultivation on a synthetic medium
m: an energy maintenance coefficient, the rate of substrate consumption for maintaining viability of one biomass unit per a unit of time
Xp: a maximum biomass concentration, when all the energy generated during cultivation is consumed for cell viability maintenance
XLim: biomass concentration in the end of exponential growth phase and beginning of growth inhibition phase
Xst: concentration of the biomass of zero age cells (stable), the content of “resting” cells
Xdiv: concentration of proliferation biomass
tLim: time of exponential growth phase termination
XLimst: concentration of stable cells at the end of exponential growth phase
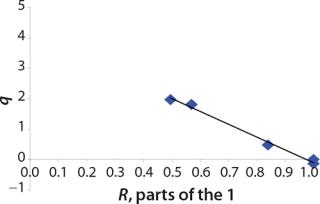
R: ratio of Xst to biomass X, relative content of st
able cells in the biomass, synchronization degree
XI: initial biomass concentration in LGP corresponding the beginning of population structuring;
Xfinal: final biomass concentration at which R = 1 (when energy consumption is limited)
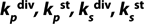
PLim: metabolite concentration at the end of LGP and beginning of GIP;
P0: metabolite concentration in LGP when biomass structuring occurs at X = XI.
Another important point is that the age structure (as revealed from our study) may change over time, not arbitrarily, but according to certain laws. Such patterns, which we have found, are the essence of the structured model that we proposed earlier for the optimization of the growth of cell populations.
It is known that stoichiometric coefficients of biochemical reactions are determined by the methods of general chemistry and biochemistry. We have accurately determined the stoichiometric coefficients of the biochemical reaction of glucose fermentation by Escherichia coli according to the structured model we proposed previously. In that case, we used experimental data from an article by Song et al (4).
Our objective for the analysis presented here is to demonstrate the possibility of describing the data obtained by Song et al. using the model shown by Klykov at al. (3, 4). We also show the possibility of determining the stoichiometric coefficients of glucose fermentation by the proposed model.
Materials and MethodsWe analyzed the data for E. coli microbial culture growth, glucose consumption, and metabolites formation using the data presented by Song et al. (4) according to the model by Klykov et al. (3, 5) presented above and shown here. Briefly, a description of this model is presented by the following thesis for the slow growth phase, GIP:
-
Nondividing cells Xst accumulate exponentially
-
The metabolite is synthesized by dividing cells Xdiv, but it can be destroyed by nondividing cells.
Equation 3–10:
Equation 3
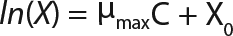
Equation 4
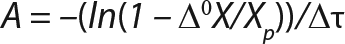
Equation 5
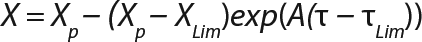
Equation 6
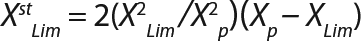
Equation 7
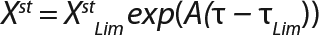
Equation 8
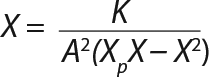
Equation 9
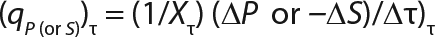
Equation 10
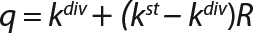
Determination of Kinetic Parameters of the Model for Biomass: In the article by Song et al. (4), Figure 6A shows anaerobic growth of E.coli glucose use as an energy substrate and biosynthesis of various metabolites. We have analyzed those processes for the proposed model of biomass growth and metabolite synthesis (3, 5). Our analysis data are presented in the following sections.
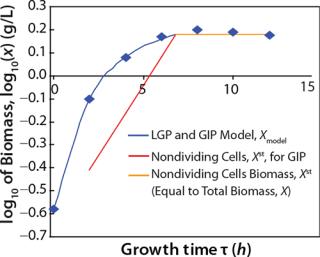
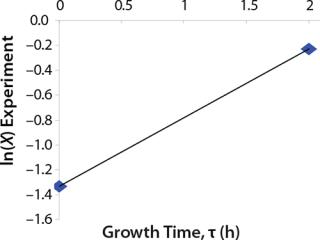
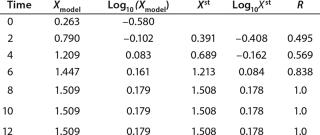
Determination of Maximum Culture Specific Growth Rate µmax: We calculated the value of µmax using the dependence of natural logarithm of the biomass on time, as Figure 1 and Table 1 show. For this value, the decimal logarithm log10(X), — presented in Figure 6A of Song et al.(4) — was converted into a natural logarithm ln(X). Using the dependence of ln(X) on time τ, gives
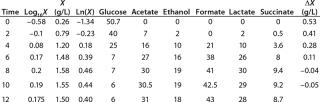
Table 1: Initial experimental biomass and metabolite data.
Despite the fact that the initial (exponential) growth phase is represented by only two points, it did not play a negative role for the entire analysis in general. In Klykov et al. (3), Figure 2 shows another method of determining µmax (Table 1).
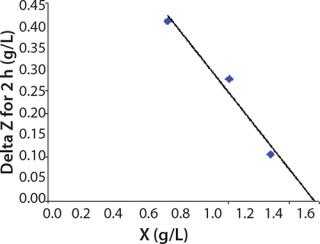
Determination of CoefficientsA,XLim, τLim,X, and Δ0X (Ordinates ofFigure 4Line) Assuming X= 0: Coefficient A is calculated after the construction of dependence ΔX2h= f(X), ΔX2h describes biomass X concentration changes during equal time intervals Δτ= 2 h. Values ΔX2h taken from Table 1 are presented in Figure 3. In that figure, the line crosses the x-axis at a value of X = Xp, and the y-axis at a value of X = 0 and ΔX = Δ0X. Figure 3 line has a slope angle equal to –0.432/h. That value represents the ratio of Δ0X/Xp, which is substituted into
Usually, the values XLim and τLim are determined in accordance with Figure 2 (3) and represent the points of intersection of exponential and straight lines for GIP. In our analysis, we used values corresponding to the transition (inflection) of biomass common logarithm line (Figure 1) at X = 0.76 g/L. Table 2 shows the parameters A, XLim, τLim, and Xp as well as the parameters for the structured model.
Table 2: Biomass
Calculation of Parameters for the Structured Model: Using
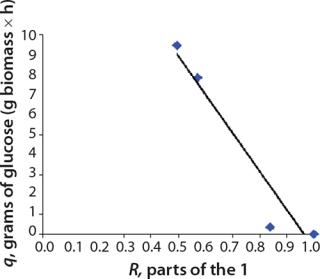
Tables 2 and 3 show the model parameters and the calculated kinetic parameters of the biomass. Figure 2 shows that by the end of growth, the biomass — as calculated by our model at hour 6.773 — reaches stationary phase. Consequently, R amounts to 1 (Table 2 and 3) and does not change further. We then calculated the dynamics of substrate consumption and product synthesis.
Table 3: Model biomass data
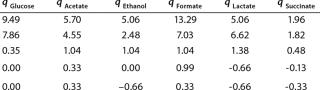
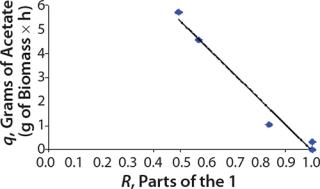
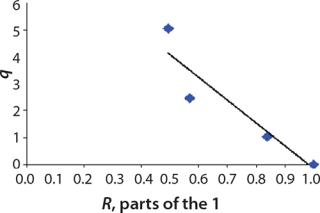
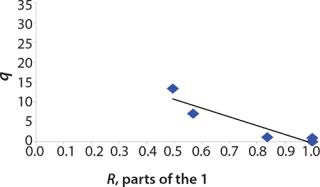
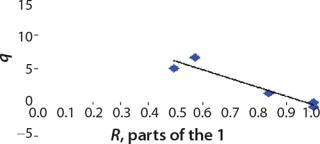
Determination of Model Parameters for Substrate Use and Metabolite Synthesis: We calculated the specific rate q substrate use and synthsis of metabolites. The parameter q for the control points (Table 3) were calculated using
Table 4: Specific synthesis rates, q
Calculation of the Rate Constants of Substrate Utilization and Synthesis of Metabolites,kdiv: Under the proposed model of substrate utilization and synthesis of metabolites, these processes take place with different specific rates of synthesis (utilization) and degradation, kdiv and kst, which correspond to dividing cells and nondividing cells of zero age, respectively. It is assumed that any process of metabolites synthesis or substrate use may be
accompanied by the opposite process of metabolite degradation or uncontrolled reduction of substrate use rate. For example, when dividing cells are produced from a metabolite substrate, nondividing cells can in turn use it as a substrate for their biosynthesis. The kinetics of such processes, as shown previously, is well described by equations with constants kdiv and kst for different cell groups Xdiv and Xst.
We calcualted the constants of synthesis and utilization according to Equation 22 in an article by Klykov et al. (3), shown as
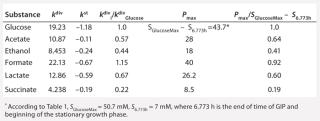
Table 5: Rate constants for various metabolites; relationship between these constants and the ratio between maximum concentrations of metabolites and glucose
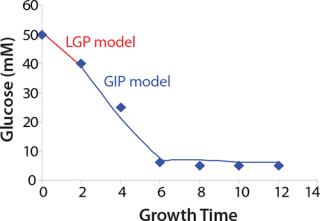
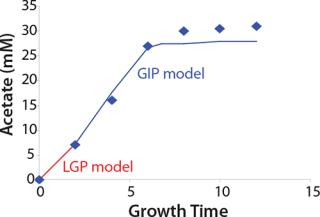
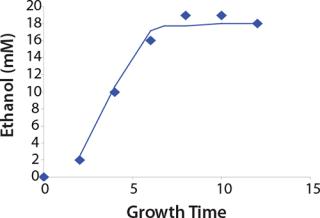
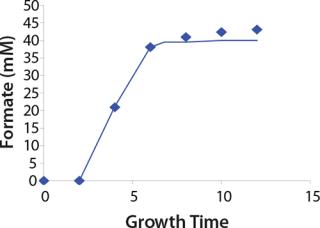
Construction of Model Curves for the Substrate (Glucose) and Metabolites: Using the above-defined constants for the models, we designed dynamic models of glucose and metabolites according to Equations 19 and 21 in Klykov et al. (3) (Figures (101112131415).
For glucose and acetate, we also calculated the process curve portions — which correspond to the exponential growth phase of the biomass (LGP). At the same time, we used constants determined from the fermentation time ranging from 2 to 6.773 hours of growth — the GIP phase. As shown in the figures, all the curves comply with experimental data, which gives evidence in favor of the proposed model.
Determination of Stoichiometric Coefficients for Glucose Fermentation According to Constants kdivand kstfor Glucose and Metabolites:Table 5 shows the constants of metabolite synthesis and glucose utilization, kdivi. Constants for the opposite process kdivi in absolute value are much smaller than kdivi, and account for only a few percents of the value kdivi. Apparently, kdivi = 0, and their nonzero value (Table 5) is due to the relative inaccuracy in the determination of the initial data.
Relations kdivi/kdivGlucose are stoichiometric coefficients characterizing a quantity of metabolites produced by one mole of the used glucose. Table 5, right columns, shows the maximum values of concentrations of the produced metabolites, Pmax, and their ratios to the amount of the consumed glucose, SGlucoseMax – S6.773h. The ratios of those variables, Pmax/(SGlucoseMax – S6.773h), are the stoichiometric coefficients, provided that kdivi = 0. That is, there are no interconversions of metabolites (not glucose) in the reactions.
As Table 5 shows, the values kdivi/kdivGlucose and Pmax/(SGlucoseMax – S6.773h) are practically identical. That gives evidence in favor of the assumption that kdivi = 0. That is also confirmed by the data in Table 6, which deals with the material balance for the individual atoms of carbon, hydrogen, and oxygen for biochemical reactions, described as follows:
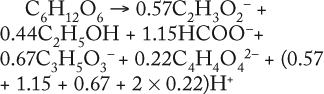
To prepare the material balance of glucose use, we multiplied the ratios of constants kdivi/kdivGlucose (Table 5), as stoichiometric coefficients by the corresponding coefficients for individual chemical elements of each individual metabolite shown in Table 5. We summed the values obtained and compared those results with the corresponding values for glucose (the middle part of Table 6). As can be seen, the material balances for the chemical elements of glucose in the left side of the biochemical reaction, and the amount of metabolites in the right side of it practically coincide with the preliminary calculation.
Benefits of the Proposed ModelOur proposed structured deterministic model describes the kinetics of biomass growth. Metabolism is not worse than other models, but rather provides a simpler mathematical approach. Our model allows for analyzing the chemical stoichiometry of biological processes. Such analysis provides for the economic calculation of the industrial processes of cell culture and a real forecast of the parameters.
Author Details
Sergey P. Klykov, PhD (
REFERENCES
//iopscience.iop.org/1758-5090/3/4/045006 3:045006.