
Figure 1: Schematic diagram of the Ambr analysis module showing the BioPAT Spectro platform fitted to an Ambr 250 high-throughput system
Raman spectroscopy is used widely in biomanufacturing as a process analytical technology (PAT) for monitoring analytes such as glucose and lactate (1). Predictive Raman models also can be used to control glucose concentration in cell cultures (2). The technique is becoming more popular for pilot- and manufacturing-scale bioreactors, but it only recently has been studied with minibioreactors for measuring analytes and producing predictive Raman models for feedback control (3) thanks to advances in integrated technology for automating sampling, analysis, and data management.
Raman-based process control relies on developing calibration models that correlate a spectral signal with an analyte or other measurements, generally from benchtop- to production-scale bioreactors. To build such a model requires measuring multiple analytes, ideally in a design of experiments (DoE) approach to cover as many expected process variations as possible by design rather than by chance. Thus, producing a predictive model can be time-consuming and costly. For example, in one study 37 bioprocess runs (from 2  L to 5,000 L) generated hundreds of data points (4). Because such a study at commercial scale is expensive in terms of media, reagents, and staff time, predictive Raman models are built by monitoring multiple production runs for variations or waiting for out-of-specification (OoS) batches to occur. Planned process variations usually cannot run in larger-scale bioreactors because of cost and waste: A manufactured batch of product must be discarded if the standard operating procedure (SOP) is not followed.
Using real-time Raman spectroscopy in minibioreactors such as an Ambr 250 bioreactor to mimic manufacturing-scale cell culture (5) makes sense because it enables cost-effective experimental design with induced variations and efficient model building. Because one run can generate data from 24–48 different bioreactor conditions or process trajectories, the design space can be much larger than one provided by running hundreds of production batches with random variations. That allows for acquisition of statistically relevant data that help users build lower-cost predictive Raman models rapidly.
Here we discuss the physical and data integration of Raman spectroscopy with Ambr bioreactors using a BioPAT Spectro spectroscopy platform. This integration enables real-time Raman spectroscopy measurement of bioreactor samples. The data generated can be analyzed by SIMCA 16 multivariate data analysis (MVDA) software to build a predictive model for glucose in a typical cell culture (SIMCA = soft independent modeling by class analogy). We then demonstrate that this predictive Raman model can be used for real-time estimation of glucose concentration in new bioreactor samples, enabling feedback control of glucose concentration in minibioreactors.
Raman Spectroscopy Integration
Adapting Ambr 15 cell-culture and Ambr 250 high-throughput systems for integrated Raman spectroscopy required connecting a user-replaceable Raman flow cell to a sample cup. A waste bottle needed to be incorporated into the Ambr analysis module, which can be fitted to either Ambr system. The flow cell is connected to a BioPAT Spectro optical probe compatible with Raman spectrometers (Kaiser Optical Systems and Tornado Spectral Systems). Samples are taken automatically from each minibioreactor (160 μL from the Ambr 15 bioreactor and 200 μL from the Ambr 250 bioreactor) by a liquid handler and released into the sample cup of the Ambr analysis module. The sample is transferred using a syringe pump to the flow cell for Raman measurement by a fiber optical probe, which connects the BioPAT Spectro flow cell to a spectrometer. After Raman measurement, the sample is discarded to waste, and a cleaning cycle is performed (using standard Ambr AM washing liquids) to prevent sample carryover.
For spiking samples, a cell culture sample is released into a well plate, and a single analyte stock solution is added by the liquid handler. Both are mixed in the well plate by pipette aspiration and dispensing, followed by sample transfer to a sample cup and delivery to the flow cell for measurement. The cycle time is ~10–15 minutes per sample. Ambr software automatically collates all key data from the Ambr bioreactors, Raman probe, and other analyzers integrated with the Ambr system. The data are merged for model construction and exported as a CSV file for statistical analysis and model building using SIMCA software.
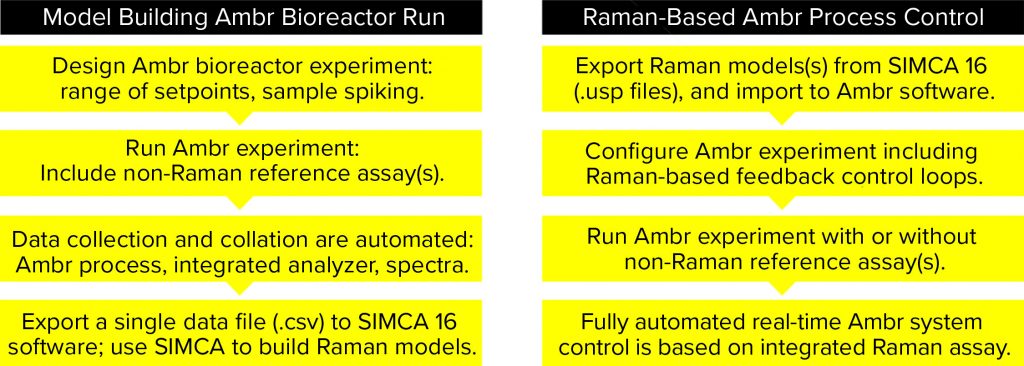
Figure 2: How to build Raman models in Ambr minibioreactors and how to use models for process control
Proof-of-Concept Process Run
To produce statistically relevant data for robust Raman model building, data were taken from 16 Ambr 250 system minibioreactors that were run using a Cellca Chinese hamster ovary (CHO) cell line, media, and process expressing a monoclonal antibody (MAb). Culture samples (0.5Â mL) were analyzed daily from the model(s) using an integrated BioProfile Flex2 automated cell culture analyzer (Nova Biomedical Corporation) as a reference standard to monitor analytes including glucose and viable cell concentration (VCC).
Raman spectra were acquired to correspond as closely as possible to the at-line reference measurements from bioreactor samples (200 µL) both before and after they were spiked with one low concentration (7.1 g/L) and one high concentration (33.3 g/L) of stock glucose solutions (spiking volume 20–60 µL). Spiked solutions were mixed in a microwell plate before analysis. From the bioreactors, 35 spiked samples were collected and measured with a RamanRXn2 hybrid in situ analyzer (Kaiser Optical Systems) as well as the reference BioProfile Flex2 analyzer. The sampling regimen consisted of one reference measurement per vessel followed by acquisition of Raman spectra from an unspiked sample and a spiked sample. Data were collated automatically in the Ambr software and exported as a CSV file into SIMCA 16 MVDA software for model building. Figure 2 summarizes Raman model building and process control.
Raman Model Building
An orthogonal partial least squares (OPLS) regression model was generated with SIMCA MVDA software using unspiked and spiked Raman spectra and corresponding glucose measurements. The results show good correlation between the observed (reference BioProfile Flex2 data) and Raman predictions for glucose in an unspiked cell culture. That demonstrates that the unspiked glucose Raman model (Figure 3, left) is a good fit with an R2 value close to 1, and low prediction errors RMSEE/RMSECV (root mean square error of estimation and root mean square error of cross validation) of 0.26 and 0.28 g/L, respectively. Those figures correspond well to the literature (6) showing comparable values using Raman spectroscopy for glucose in unspiked samples cultured in minibioreactors, indicating that this Raman model could be used to predict glucose concentrations with a similar accuracy to observed concentrations.

Figure 3: Comparing reference and Raman model plots of glucose in unspiked (left, n ~250) and spiked (right, n ~280) cell culture samples from Ambr 250 minibioreactors (RMSEE = root mean square error of estimation; RMSECV = root mean square error of cross validation)
Results from the spiked Raman model (Figure 3, right) provide a proof of methodology for specific spiking in the Ambr bioreactor. Spiking successfully extended the calibration range of the glucose model, an important aspect of robust model generation involving other analytes such as lactate, for which typical process ranges are much smaller.
Raman-Based Process Control
Before the feedback control demonstration run, a Raman glucose model that had been built in SIMCA 16 was loaded into the Ambr software. That was used for real-time glucose monitoring throughout a 12-day process alongside reference glucose measurements using an integrated BioProfile Flex2. To determine the potential of using Raman-predicted glucose values for process control, glucose was maintained at 5 g/L from day 6 onward using either Raman predictions (n = 4) or BioProfile Flex2 measurements (n = 4). The results show that Raman glucose predictions align very closely with reference analyzer values (Figure 4, left) and when used to control glucose, lead to comparable VCC profiles (Figure 4, right). This indicates that the BioPAT Spectro analyzer in an Ambr system enables fully automated glucose control capabilities similar to those of other analytical techniques and demonstrates the ability to control a cell culture process using only a glucose Raman model.
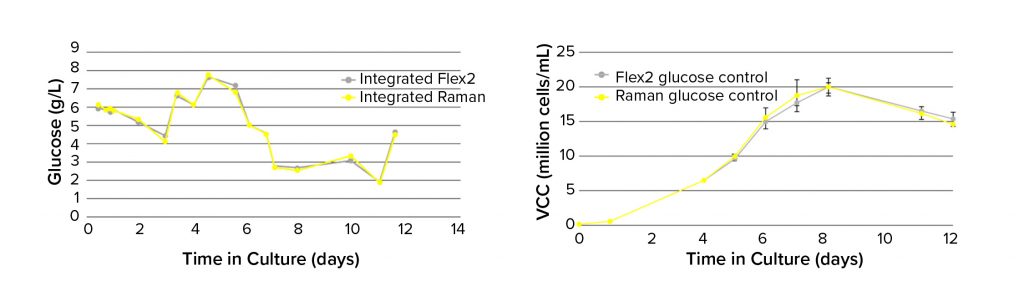
Figure 4: Comparing reference (BioProfile Flex2 analyzer) and Raman model plots of glucose concentration (left, n = 1) and VCC (right, n = 4) in Ambr 250 minibioreactors.
Those results are promising. BioPAT Spectro analyzers in minibioreactors use Raman probes and flow-cell optical paths similar to those used for on-line Raman spectroscopy monitoring in Biostat STR single-use pilot and commercial-scale bioreactors (50–2,000 L). So the next step is to transfer Raman models built based on small-scale data to those larger bioreactors to demonstrate the scalability of Ambr-derived Raman models.
A Robust Glucose Model
We have described integration of Raman spectroscopy with Ambr minibioreactor systems using BioPAT Spectro probe technology. We have demonstrated in a proof-of-concept study using a MAb-expressing cell line cultured in Ambr 250 bioreactors that glucose measurements from Raman spectra generated by BioPAT Spectro show good correlation with observed reference data from a BioProfile Flex2 integrated analyzer.
Based on the capabilities of automated spectral acquisition and spiking, as well as the automatic alignment of reference data, we have shown that an Ambr 250 high-throughput system can generate a robust Raman glucose model. The multiparallel nature of the Ambr system facilitates DoE, potentially enabling robust models for glucose and other analytes of interest.
We also have demonstrated from a process run in Ambr 250 minibioreactors that MAb-expressing cell cultures controlled by a predictive Raman model for glucose have similar glucose and VCC profiles compared with reference data. Ambr Raman prediction models have robust built-in process variations that can be transferred and exploited in large-scale production bioreactors with significantly reduced calibration effort.That gives them utility in reducing the timelines and costs in manufacturing biologics.
References
1 Buckley K, Ryder AG. Applications of Raman Spectroscopy in Biopharmaceutical Manufacturing: A Short Review. Appl. Spectrosc. 71(6) 2017: 1085-1116; https://doi.org/0.1177/0003702817703270.
2 Berry B, et al. Cross-Scale Predictive Modeling of CHO Cell Culture Growth and Metabolites Using Raman Spectroscopy and Multivariate Analysis. Biotechnol. Prog. 31(2) 2015: 566–577; https://doi.org/10.1002/btpr.2035.
3 Rowland-Jones RC, Jaques C. At-Line Raman Spectroscopy and Design of Experiments for Robust Monitoring and Control of Miniature Bioreactor Cultures. Biotechnol Prog. 35(2) 2019: e2740; https://doi.org/10.1002/btpr.2740.
4 Li B, et al. Performance Monitoring of a Mammalian Cell-Based Bioprocess Using Raman Spectroscopy. Anal. Chim. Acta 796, 2013: 84–91; https://doi.org/10.1016/j.aca.2013.07.058,
5 Manahan M, et al. Scale-Down Model Qualification of Ambr® 250 High-Throughput Mini-Bioreactor System for Two Commercial-Scale MAb Processes. Biotechnol. Prog. 2019: e2870; https://doi.org/10.1002/btpr.2870.
6 Rowland-Jones R.C, et al. Comparison of Spectroscopy Technologies for Improved Mmonitoring of Cell Culture Processes in Miniature Bioreactors. Biotechnol. Prog. 33(2) 2017: 337–346; https://doi.org/10.1002/btpr.2459.
Corresponding author Michael Sibley is a development scientist at Sartorius; 44-1763-227200; michael.sibley@sartorius.com; www.sartorius.com/biopat-spectro.
