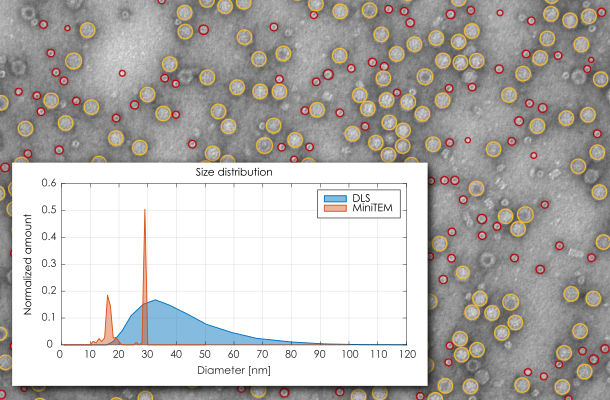
Characterization and analysis of subvisible particles of biological origin can be challenging or give insufficient information. In this study Adeno associated virus (AAV) particles contaminated by host cell proteasomes are analysed with direct versus indirect methods performed in a standard laboratory setting to reveal the difference in performance and quality of the information obtained. The focus is on the ability to detect the AAV particles of interest and distinguish them from the contaminating proteasomes. The compact, tabletop MiniTEM system enables automatic sample screening and imaging together with direct measurements of particle size and particle classification. This method is compared with the indirect methods Dynamic Light Scattering (DLS) and Nanoparticle Tracking Analysis (NTA) measurements using Zetasizer Nano and NanoSight NS300 respectively. Both the DLS and NTA methods measure the Brownian motion of the particles or molecules in suspension and use the Stokes-Einstein equation to give the particle size distribution.
Monday November 17, 2025