
Photo 1: The ambr250 scale-down bioreactor system controls 12 or 24 disposable small-scale
bioreactors (100–250 mL working volume) and offers parallel processing and evaluation of multiple experiments while maintaining the characteristics of a larger scale bioreactor.
Design of experiments (DoE) is one of the most valuable techniques for organized and efficient planning, execution, and statistical evaluation of experiments. Although a DoE investigation can be completed using several runs in one bioreactor, small-scale bioreactor systems designed for parallel operation (such as the ambr15 or ambr250 systems) provide the optimal basis to economically realize a series of experiments. Because of the multitude of interdependent parameters involved in applications such as cell line development, culture media screening, and the optimization of bioreactor operating parameters, DoE evolved as an essential and indispensable method to create process knowledge. It has improved speed of development and has helped define manufacturing processes for high-quality products.
Design of Experiments: The Efficient Strategy
In pharmaceutical bioprocessing, decreased development time and production costs are key objectives. Within this context, the combined application of process analytical technology (PAT) tools and reliable flexible bioprocess equipment is the basis for an efficient optimization of existing production processes and the development of new ones. Spearheaded by the FDA initiative to use PAT for greater control and process understanding, statistical DoE methods are extensively applied to look for the best process conditions while reducing expensive and time-consuming experiments to a minimum (1).
The concept of DoE is to vary process parameters simultaneously over a set of planned experiments and then interpret the results by means of a proven mathematical model. This model can be used subsequently for interpretation, prediction, and optimization, which allows for greater understanding of processes. By contrast to changing one factor at a time, the DoE procedure delivers optimized information content by using the least number of experiments, thereby reducing development time and labour.
The improvement of production and feed media components is crucial to providing an environment optimal for growth of a used recombinant cell line and formation of a functionally active therapeutic protein (2). Another key target is the optimization of basic process parameters such as temperature, pH, and dissolved oxygen (3). Often, optimization of culture media and state variables are combined, whereas DoE studies can significantly facilitate such tasks (1).
Transition from Medium and Feed Development to Process Optimization
Apart from the productivity of a specific cell line, the culture medium has an important impact on both yield and quantity of therapeutic protein and monoclonal antibody produced. The cells require a combination of macro- and micronutrients as sugars, trace salts, vitamins, amino acids, and other components to support cell growth and protein production. Because of the diverse nutritional requirements that are unique to every cell and the large number of medium factors, small-scale systems — with their ability to perform a large number of experiments in parallel — are needed to unlock development bottlenecks.

Photo 2: The BIOSTAT B-DCU II system is designed for advanced process optimization and characterization and is available with working volumes of 0.5–10 L; it features independent process control for up to six culture vessels
Historically, media and feed formulations are derived through numerous empirical tests by changing one factor at a time (4). This methodology is simple and convenient, but it ignores interactions between components, can miss the optimum completely, and is fairly time-consuming. A superior approach to tackle the problem of media design and shorten the time needed for medium development is to use high-throughput cell culture scale-down systems in combination with statistical DoE approaches.
In addition to improving the medium composition, optimization of the feed-control strategy can be beneficial for fed-batch processes. The practical approach depends on the infrastructure of the bioreactor system itself and the cultivated cell line. Whereas a classical fed-batch with continuous feeding strategies has been proven to be successful for Escherichia coli and Pichia pastoris cultivations, the method of adding a stepwise bolus of feed solution to a production bioreactor is most widely used in industry for mammalian cell culture because of its simplicity and scalability (5, 6).
The new ambr250 workstation with 12 or 24 single-use stirred tank reactors (TAP Biosystems, a Sartorius company) represents an efficient, high-throughput, scale-down model for initial process development of microbial fermentation or cell culture. The system provides the capability to continuously monitor and control critical parameters in real-time as well as extended feed and sampling options to enable a more effective scale-up and to demonstrate equivalent process performance in comparison to laboratory and pilot scale (7–9).
The automated workstation features an integrated liquid handler, which can remove the cap of a single-use vessel to enable initial media load, seeding, sampling, and bolus additions. Culture or feed medium can be transferred preformulated by the pipetting module, or the system can be used to automatically make up media from different components using imported experimental designs. Besides single bolus additions (10 μL to 10 mL), integrated pumps allow continuous feeds of linear or exponential profile ranging from 20 nL/h to 20 mL/h.
Once the most prominent medium components and their optimal ranges are identified, the process can be transferred to a larger laboratory-scale system for experimental verification of the identified optimum to test scalability and to continue with optimizing the bioreactor’s operating parameters as well as process characterization studies.
The BIOSTAT B-DCU II benchtop bioreactor (Sartorius Stedim Biotech) is specifically designed for laboratory-scale parallel operations of up to six multiuse or single-use culture vessels with one control tower. It is used widely in the industry for process optimization and characterization (10). Each vessel has independent process controls with a wide range of measurement and automation features. Systems are preconfigured for immediate use, and they can be used for microbial fermentation or cell culture packages.
Optimization of Recombinant Protein Expression: A Case Study
Within process development divisions, the goal is to identify key parameters that maximize target yield and product quality within process parameter ranges that can be reliably attained at pilot and production scale. In the following case study, a BIOSTAT Q plus six-fold system was used in conjunction with DoE to optimize recombinant protein expression in two E. coli BL21 (DE3) strains.
E. coli is one of the most used prokaryotic organisms for the production of active pharmaceutical ingredients (APIs) or parts thereof (e.g., antibody-drug conjugates, ADCs). Testing novel APIs requires the physiologically active form of the protein for preclinical toxicology studies. Although protein solubility does not necessarily correspond to active protein, fast product availability often is limited by low soluble protein yields.
Growth rate, cultivation temperature, and IPTG inducer concentration were investigated for each E. coli strain. The effect of each factor and strain on the space–time yield of soluble protein and inclusion bodies was used for process evaluation. Expressed protein was tagged with a green fluorescent protein, allowing for simple and rapid quantification of protein expression with a common fluorescence reader.
For advanced process monitoring and control, the BIOSTAT Q plus system was equipped with O2 and CO2 off-gas analysers. Values from those sensors and with the BioPAT MFCS/win bioprocess software allowed for online calculation of cell-specific growth rates as well as data storage and supervisory control. The DoE software BioPAT MODDE was used for experiment definition, statistical evaluation of raw data, and construction of an easy to interpret model.
The Workflow to Success
An experimental design was created to screen for variables that would have the highest effect on the space–time yield. Given the known dependence of growth rate on temperature, the DoE software was used to design a set of experiments with varying levels of each factor. The final experimental plan included four runs on a centered level to determine the variability within the overall system.
Using the BIOSTAT Q plus system, it was possible to screen each E. coli strain in as few as 12 runs to identify the most promising strain for further studies. In addition, the inducer concentration was shown to have no significant effect on product yield, so that the lowest investigated concentration could be used for further studies. Once the initial screening procedure was completed, higher levels for growth rate were tested in combination with lower temperature set-points.
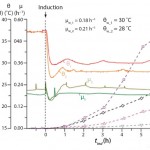
Figure 1: Comparison of two protein production phases with different factor settings 1 and 2; Sk = fluorescence signal of internal soluble (k = sol) and insoluble (k = IB) protein fraction; θ = cultivation temperature; μ = online observed cell specific growth rate
Figure 1 shows the optimization potential of the DoE approach while simultaneously varying factor conditions. Using the lowest inducer concentration, different set-points for growth rate (μw) and cultivation temperature (θw) resulted in a higher soluble and insoluble protein yield for the high/low factor level combination subscripted with 2.
Through this set of optimization experiments and continued interpretation within the DoE software, the process could be further understood finally leading to a substantial increase of soluble protein concentration. Highly predictive models gave a reliable direction to obtain high space–time yields at low temperature in combination with high growth rate. Figure 2 displays the predicted space–time yields as a response surface spanned by the two factors.
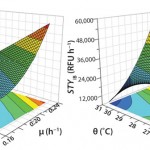
Figure 2: Response surface plots for soluble (left) and insoluble (right) space-time yield; STYk = soluble (k = sol) and insoluble (k = IB) space-time yield; θ = cultivation temperature; μ = cell-specific growth rate
The final test in this experiment was a robustness trial, testing parameters for the maximum allowable process parameter ranges without influencing product quality. Using only six experimental conditions, a safe operating range could be confirmed in which the desired space–time yields were achieved. The design space tools provided by the BioPAT MODDE software can be used to visualize an operating range for the investigated process parameters in a unique probability contour plot considering risk analysis specifications. That guides engineers in determining how likely it is that their experiments will truly identify the most reliable operating parameter ranges.
Conclusions
High-throughput cell-culture scale-down bioreactor systems such as the ambr 15 and the ambr 250 systems can increase efficiency and effectiveness of screening, process development and process characterization exercises. They offer the advantage that a significant number of experiments can be performed simultaneously at small culture volumes. At the same time, the culture environment and bioreactor characteristics of small-scale single-use stirred tank vessels provide excellent comparability to larger scale bioreactors (7–9). Leaving the traditional way of trial-and-error optimization behind, advanced DoE approaches enable fast and effective identification of critical process parameters and provide significant cost savings and reduction of development timelines.
Acknowledgments
Experimental work has been performed at the Research Center of Bioprocess Engineering and Analytical Techniques from the Hamburg University of Applied Sciences, headed by Prof. Dr-Ing. R. Luttmann.
References
1 Mandenius CF, Brundin A. Bioprocess Optimization Using Design-of-Experiments Methodology. Biotechnol. Prog. 24, 2008: 1191– 1203.
2 Dong J, et al. Evaluation and Optimization of Hepatocyte Culture Media Factors By Design of Experiments (DoE) Methodology. Cytotechnol. 57, 2008: 251–261.
3 Fricke J, et al. Designing a Fully Automated Multibioreactor Plant for Fast DoE Optimization of Pharmaceutical Protein Production. Biotechnol. J. 8, 2013: 738–747.
4 Weuster-Botz D. Experimental Design for Fermentation Media Development: Statistical Design or Global Random Search? J. Biosci. Bioeng. 90(5) 2000: 473–483.
5 Shiloach J, Fass R. Growing E. coli to High Cell Density: A Historical Perspective on Method Development. Biotechnol Adv. 23(5) 2005: 345–357.
6 Li F, et al. Cell Culture Processes for Monoclonal Antibody Production. mAbs 2(5) 2010: 466–479.
7 Hsu WT, et al. Advanced Microscale Bioreactor System: A Representative Scale- Down Model for Benchtop Bioreactors. Cytotechnol. 64, 2012: 667–678.
8 Bareither R, et al. Automated Disposable Small-Scale Reactor for High Throughput Bioprocess Development: A Proof of Concept Study. Biotechnol Bioeng. 110(12) 2013: 3126–3138.
9 Ngibuini M. Automated Mini Bioreactor Technology for Optimizing Early Process Development in Fermentation and Bioprocessing. BioProcess Int. 12(8) 2014: S3– S6.
10 Tsang VL, et al. Development of a Scale Down Cell Culture Model Using Multivariate Analysis as Qualification Tool. Biotechnol. Prog. 30(1) 2013: 152–160.
Corresponding author Andree Ellert is product manager, bioprocess software at Sartorius Stedim Biotech GmbH (andree. ellert@sartorius-stedim.com), and Conny Vikström is senior application specialist, product manager MODDE at Umetrics AB, 46 90-184849; conny.vikstrom@umetrics.com.
