Typically a concern with cereal grains, mycotoxins are produced by filamentous fungi under specific conditions, as shown here on damp buckwheat. Animals that consume contaminated feed can present detectable mycotoxin levels in their milk (https://www.istockphoto.com)
Sevenfact eptacog beta is a new recombinant human factor VIIa (rFVIIa) developed by LFB SA in Les Ulis, France, as a bypassing agent (BPA) for treatment and control of bleeding in people with hemophilia A and B and inhibitors (1, 2). The product was approved for use in adults and adolescents by the US Food and Drug Administration (FDA) in April 2020 (3). It is expressed in the milk of transgenic rabbits and purified through a multistep process using both filtration and chromatography. The source material (transgenic rabbit milk) naturally contains a microbiological load. To manage that, the collection process is controlled carefully, resulting in a historical average bioburden of 1,800 CFU/mL (n = 116). That is well below the prepasteurization standard of ≤100,000 CFU/mL for US grade A cow’s milk (4). The Sevenfact clarification and purification process is designed to clear bacterial load rapidly from the source material. Bacterial load and endotoxins are monitored throughout the process to confirm rapid clearance of the native flora and demonstrate process control.
In addition to bacterial load is the potential for a fungal load. With the source-material collection process, it is unavoidable to have occasional fungal presence from animals and their environment. To manage that, total yeasts and molds are monitored throughout the manufacturing process, and they have been shown to clear quickly early in that process. However, it is not known whether potentially toxic by-products of fungal growth (mycotoxins) accompany the presence of detected molds because such toxins cannot be measured easily or monitored in process streams.
Mycotoxins are toxic secondary metabolites produced by some species of filamentous fungi (molds) including Aspergillus, Penicillium, and Fusarium (5). Typically a concern for storage of cereal grains, mycotoxins are produced under specific conditions of temperature and pH, with the availability of simple carbohydrates for active fungal growth that often can be found in such applications (5). Animals that consume contaminated feeds can present detectable levels of mycotoxins (primarily aflatoxin M1) in their milk. Accordingly, the FDA has set an aflatoxin action limit of 0.5 ppb in milk (6) and a maximum level for it in the feedstock of lactating dairy animals of 20 ppb (7). Vendors providing rabbit feed to LFB are selected with this aspect taken into consideration, with all feed tested for aflatoxins to ensure mycotoxin safety and eliminate diet as a potential source for mycotoxin exposure.
 Our aim with the research reported herein was to understand better the mycotoxin risk for Sevenfact drug substance by evaluating the potential for fungal growth and mycotoxin generation in transgenic rabbit milk. In addition, we wanted to determine the mycotoxin clearance capability of the Sevenfact purification process. To evaluate the potential for fungal growth and mycotoxin generation, LFB examined its transgenic rabbit milk over time after spiking it with a quantitative amount of a commercially available fungal standard. For clearance evaluation, relevant commercially available mycotoxins were spiked into process fractions from a scaled-down process representation to determine the ability of our purification process to remove mycotoxins that may be present in the Sevenfact source material. The clearance potential of each process step was evaluated to determine the mycotoxin clearance of the total process. To calculate the risk to patients through a theoretical dosing exercise, we summed the individual step clearances and estimated the safety factor in excess of published mammalian toxicity levels (5).
Our aim with the research reported herein was to understand better the mycotoxin risk for Sevenfact drug substance by evaluating the potential for fungal growth and mycotoxin generation in transgenic rabbit milk. In addition, we wanted to determine the mycotoxin clearance capability of the Sevenfact purification process. To evaluate the potential for fungal growth and mycotoxin generation, LFB examined its transgenic rabbit milk over time after spiking it with a quantitative amount of a commercially available fungal standard. For clearance evaluation, relevant commercially available mycotoxins were spiked into process fractions from a scaled-down process representation to determine the ability of our purification process to remove mycotoxins that may be present in the Sevenfact source material. The clearance potential of each process step was evaluated to determine the mycotoxin clearance of the total process. To calculate the risk to patients through a theoretical dosing exercise, we summed the individual step clearances and estimated the safety factor in excess of published mammalian toxicity levels (5).
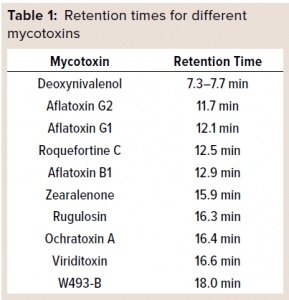
It is impossible to evaluate each of the hundreds of possible mycotoxins individually (8). Thus, we sought to classify them according to their general physiochemical properties and then apply a bracketing approach by evaluating only the worst-case examples for which commercial standards were available. To that end, LFB developed an analytical method based on high-performance liquid chromatography (HPLC) that could resolve 10 different commercially available mycotoxin standards easily. From those we chose viriditoxin (the most hydrophobic) and deoxynivalenol (DON, the most hydrophilic) for the process-clearance studies because they bracketed all the other mycotoxin standards in our hydrophobicity assay evaluations.
We considered physical size to be constant for all mycotoxins because the vast majority of them have a molecular weight of <1,000 g/mol (5), which is at least 50Ă— smaller than the eptacog beta recombinant human factor VIIa molecule. We also considered ionic interactions of mycotoxins to be constant because most of them are not charged and generally are only slightly soluble in water. Therefore, we considered their variability in hydrophobicity to be the largest differentiator for our study and chose mycotoxins that bracketed the entire observed hydrophobicity range.
Materials and Methods
Mycotoxin Preparation: DON from Sigma-Aldrich, rugulosin from Abcam, and viriditoxin from Adipogen were dissolved in ultrapure dimethyl sulfoxide (DMSO) from VWR to a working concentration of 2.5 mg/mL and stored at –20 °C until use. Those mycotoxin stock solutions were used both in the HPLC assay and as process spiking material. Taken together, they represent a full range of hydrophobicity based on retention times of 10 different mycotoxins evaluated with the C18 assay described below.
HPLC Assay: Mycotoxins were separated on a 4.6 mm × 150 mm, 5-µm Gemini C18 column (Phenomenex). Mobile phases A and B contained 5 mM ammonium acetate with 1% acetic acid and 10% or 97% methanol, respectively. We adapted a linear gradient from a Phenomenex technical bulletin (9), with detection at both 260 nm and 280 nm.
Ten mycotoxins were evaluated during assay development: aflatoxins B1, G1, and G2; DON, ochratoxin A, roquefortine C, rugulosin, viriditoxin, W493-B, and zearalenone. Table 1 lists relative retention times for each mycotoxin evaluated. Individual standard curves were generated for each mycotoxin used in the process-clearance study.
Sample Extraction Procedure: LFB collected samples of each spiked process fraction as well as the resulting process step output and stored them all at –80 °C before analysis. To each 200-µL sample, 600 µL of DMSO was added. Samples were vortexed and passed through a 10-kDa molecular-weight cut-off (MWCO) Microsep Advance polyethersulfone (PES) membrane from Pall to remove residual protein. Filtrates were loaded directly onto the C18 column without dilution. Extraction efficiency was evaluated but not considered in our study calculations because we determined clearance by direct comparison of each process step’s input and output, assuming extraction efficiency to affect each equivalently.
Milk Growth Studies: Raw and pasteurized transgenic milks were spiked with Penicillium hirsutum standard from Microbiologics. P. hirsutum is a known mycotoxin producer of roquefortine C, and the genus is isolated commonly from our transgenic rabbit colony. Spiked milk was incubated at room temperature for up to five days to allow for sufficient fungal/mold growth to occur. Samples were taken at multiple intervals and analyzed for bacteria, mold, and mycotoxins.
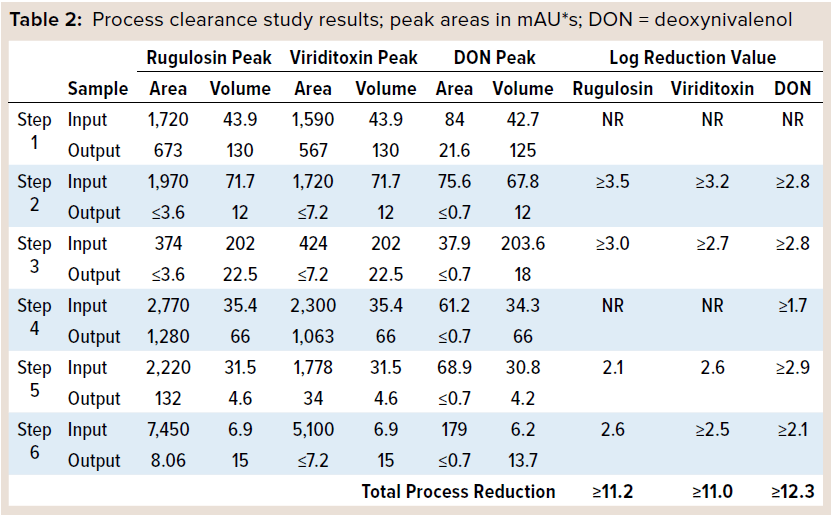
Process Scale-Down: Each process step was scaled down to minimize fraction volumes while maintaining process comparability. When possible, the process steps were performed at the same scale as in our previous process robustness studies. Six process steps were evaluated for mycotoxin clearance: (1) depth filtration, (2) affinity chromatography, (3) ultrafiltration/diafiltration (UF/DF), (4) anion-exchange chromatography, (5) mixed-mode chromatography, (6) and size-exclusion chromatography (SEC) — as numbered in Table 2.
Results
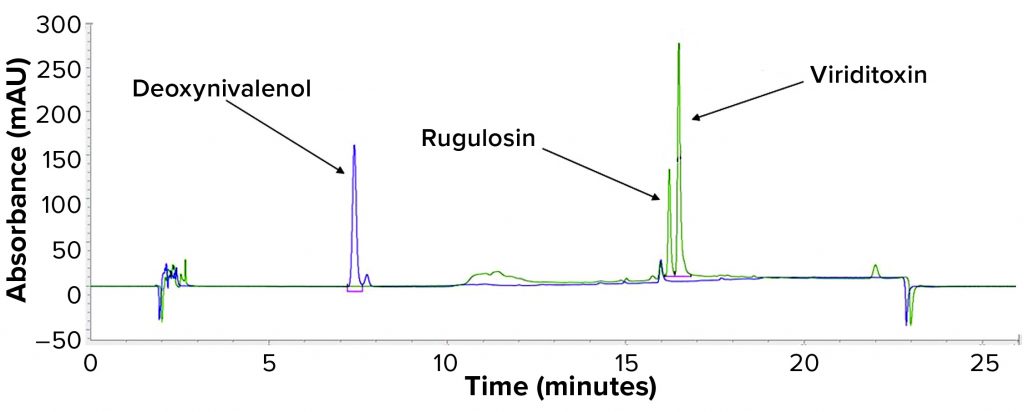
Process Clearance: To demonstrate proportionality between concentration and peak area and to determine the minimum detectable amount of each mycotoxin, we prepared a series of standards in DMSO for rugulosin, viriditoxin, and DON. Both rugulosin and viriditoxin eluted as single peaks with maximum response at 260 nm (Figure 1). DON eluted as a doublet with peaks at 7.3 and 7.7 minutes. Maximum absorbance was at 260 nm for the 7.3-min peak and 280 nm for the 7.7-min peak, and the two were summed for quantitation purposes: DON = (7.3-min peak area at 260 nm) + (7.7-min peak area at 280 nm).
Each mycotoxin standard curve was linear (Figure 2), with significant differences in absorptivity between two mycotoxins: DON with ~20-fold lower absorption than results showed for rugulosin. We determined the limit of detection (LoD) to be 0.1 µg/mL for rugulosin and viriditoxin and 0.5 µg/mL for DON.
Next, mycotoxin standards were spiked into the input material for each process step in two separate studies. Rugulosin and viriditoxin were spiked together in the first study, with DON spiking in a separate study (DON was added later to achieve the stated hydrophobicity bracketing approach). We processed the spiked inputs and collected the output of each process step according to normal process operations. Samples of each input and output were extracted with DMSO, then proteins were removed with a 10-kDa membrane. Filtrates were loaded directly onto a C18 column and determined the resulting mycotoxin peak areas. Output samples below the LoD were loaded at larger injection volumes to increase the sensitivity of the technique by fivefold. Step clearances were determined by comparing the mycotoxin peak area times the sample volume from the input and output fractions. Table 2 reports the results as logarithmic reduction values (LRVs).
All process samples contained an interfering peak at 16.3 minutes that could not be resolved from the rugulosin peak. The interference was present in unspiked process samples that had been subjected to the extraction process. Further investigation determined that the peak was extracted by DMSO from the 10-kDa PES membrane used to remove protein before C18 analysis. We investigated alternative mycotoxin extraction methods to eliminate that DMSO–PES incompatibility issue but found no alternative that could extract the majority of the mycotoxins consistently from varying sample matrices throughout the process. Therefore, a background-subtraction technique was developed by relating the 16.3-min extract peak to another PES extract peak at 17.0 minutes, which was spatially isolated in the chromatograms.
We determined that the 16.3-min extract peak was consistently 38% of the 17.0 min peak in unspiked investigational extracts, regardless of the sample matrix. Hence, the rugulosin peak area in spiked samples could be calculated by subtracting 38% of the 17.0-min peak from the total peak area at 16.3 minutes. The remainder served as a reliable estimation of the rugulosin peak area in the presence of technique-based background peaks. Similarity between the rugulosin and viriditoxin individual process step-clearance factors (similar retention at 16.6 minutes with no interference) supported the suitability of our calculated background correction.
Growth in Milk: Once we had determined the process capabilities of mycotoxin removal, another study was executed to estimate the maximum amount of mycotoxin that could be present in the process source material (milk). From the results of this study, we could calculate potential risks to patients in a worst-case scenario of mold contamination and subsequent growth.
Raw milk was inoculated with P. hirsutum standard (a known mycotoxin producer and previously isolated genus in rabbit source material) and incubated at ambient temperature for up to five days. Samples were taken at set time points for microbial counts and mycotoxin determination. No mold growth was observed over five days: It appeared that bacterial growth had increased quickly and outcompeted the inoculated mold. Consequently, a second experiment was initiated using pasteurized milk to eliminate the native microbes and thus evaluate inoculated mold growth. The pasteurized milk was inoculated and monitored for four days, similar to the procedure for the previous study. With bacterial growth successfully eliminated, exponential mold growth was detected on the third and fourth days (Table 3).
Samples from both mold-growth studies were extracted with DMSO and analyzed in the HPLC assay as above. A series of roquefortine C (mycotoxin produced by P. hirsutum) standards also were analyzed to enable estimation of mycotoxin levels in the growth study. A background extract peak again interfered with our analysis as with rugulosin in the clearance study. So we attempted a similar background subtraction technique, but the results were not as consistent in this case — with sporadic negative values obtained. Without correction, the peak area fluctuated between ~200 and ~800 mAU*s for all test points with both raw and pasteurized milk, regardless of mold growth. We saw no peak-area differences between the raw- and pasteurized-milk studies, even though mold growth was substantial in the pasteurized milk. That suggests that mycotoxin was produced at low levels or not at all (with the same assay noise level both with and without confirmed mold growth).
Patient Safety: For the sake of patient safety, we chose a conservative starting point by presuming that the roquefortine C peak was due solely to the mycotoxin rather than sample-extraction interference. With that approach, the maximum level of roquefortine C observed in the growth study was 0.26 µg/mL. For a worst-case calculation exercise, it is presumed that all source material was contaminated to that level. Given the amount of source material required for a drug substance (DS) lot, a total of 117 mg of mycotoxin could be present in the milk batch. Each DS represents about 1,200 maximum doses for 75-kg patients, for each of whom the maximum mycotoxin dose would be 98 µg or 1.3 µg/kg. By applying the minimum process clearance value determined in this work (11 log10), we found that the mycotoxin concentration is reduced to 1.3 × 1011 µg/kg through downstream purification. By conservatively adopting the most toxic LD50 (2.8 mg/kg of viriditoxin for mice, intraperitoneal) among the mycotoxins studied, the calculated worst-case dose is 220 trillion times lower than the published mammalian LD50.
Discussion
LFB initiated these studies to estimate the impact of potential mycotoxin exposure for patients using our company’s Sevenfact recombinant clotting factor. Three mycotoxins — rugulosin, viriditoxin, and DON — were chosen to represent the full range of physiochemical properties and reasonably bracket all potential mycotoxins from different mold sources. To determine the process-step clearances, we spiked each mycotoxin into the starting material for each individual purification step. Samples were solvent-extracted and quantitated using a C18 reversed-phase (RP-HPLC) assay.
Each mycotoxin studied was cleared effectively by the six-step purification process, with minimum cumulative clearance of ≥11 LRV. Four of the six process steps were highly effective in clearing mycotoxins. The two size-based process steps — UF/DF and SEC — efficiently removed mycotoxin as expected considering the large size differential between the mycotoxins and the protein of interest. Two other process steps — affinity and mixed-mode chromatography — rely on different modes of chromatographic separation in which mycotoxins are not bound to the functional groups and thus washed away before product elution.
Concurrent with the clearance study, we also performed a mold-growth study on our source material. Raw source material was inoculated with P. hirsutum, a known mycotoxin producer of a genus that has been isolated from the transgenic rabbit colony. Mold and bacterial growth were monitored over five days. The bacterial population of the native source material grew exponentially within 24 hours and appeared to outcompete the inoculated mold, which showed little to no growth over five days at ambient temperature. We repeated the experiment using pasteurized source material to kill the native flora before inoculation. Without bacterial competition, P. hirsutum did grow in the pasteurized source material after three days of incubation at ambient temperature.
Samples from both source-material growth studies were solvent-extracted and analyzed for roquefortine C content using RP-HPLC. But an interfering peak from the solvent extraction process had coeluted with the mycotoxin, making the results unquantifiable. Because the fluctuating values obtained were qualitatively similar for both studies, even without mold growth in the raw source material and with exponential growth in the pasteurized source material, we surmised that little or no mycotoxin had been produced when the mold was actively growing by day four.
Because roquefortine C levels could not be quantitated in the growth study, we chose a conservative worst-case mycotoxin concentration from the highest peak area observed during our study to represent the interference peak combined with roquefortine C that may have coeluted. Based on that overestimation, we calculated patient risk using source-material/DS batch size, maximum dose for a 75-kg patient, and the process clearance values reported herein. Under the worst-case assumptions, the concentration of potential mycotoxins in a dose was calculated to be 220 trillion-fold lower than the published mammalian LD50 value for viriditoxin. That demonstrates robust safety relative to mycotoxins. Note that these estimates are based on source material modified to remove native flora, spiked with active mold, and held at ambient temperature for multiple days. That is not representative of normal production operations, in which the source material is frozen quickly without pasteurization and then processed on the same day that batches are thawed and pooled.
References
1 Wang M, et al. PERSEPT 1: A Phase 3 Trial of Activated Eptacog Beta for On-Demand Treatment of Haemophilia Inhibitor-Related Bleeding. Haemophilia 23(6) 2017: 832–843; https://doi.org/10.1111/hae.13301.
2 Ducore J, et al. Safety and Dose-Dependency of Eptacog Beta (Activated) in a Dose Escalation Study of Non-Bleeding Congenital Haemophilia A or B Patients, With or Without Inhibitors. Haemophilia 23(6) 2017: 844–851; https://doi.org/10.1111/hae.13357.
3 Sevenfact Package Insert. HEMA Biologics, LLC: Louisville, KY, 2020; https://sevenfact.com/Content/PDF/Sevenfact_PI.pdf.
4 Grade “A” Pasteurized Milk Ordinance. US Food and Drug Administration: Rockville, MD, 2019; https://www.fda.gov/media/140394/download.
5 Wong DT, Hamill RL. Viriditoxin Induces Swelling and ATPase By Activation of Calcium Transport in Liver Mitochondria. Biochem. Biophys. Res. Commun. 71(1) 1976: 332–338.
6 ORA. CPG Section 527.400 Whole Milk, Lowfat Milk, Skim Milk – Aflatoxin M1. US Food and Drug Administration: Rockville, MD, November 2005; https://www.fda.gov/regulatory-information/search-fda-guidance-documents/cpg-sec-527400-whole-milk-lowfat-milk-skim-milk-aflatoxin-m1.
7 CVM/ORA. FDA Section 683.100 Action Levels for Aflatoxins in Animal Food: Compliance Policy Guide. US Food and Drug Administration: Rockville, MD, March 2019; https://www.fda.gov/media/121202/download.
8 Brase S, et al. Chemistry and Biology of Mycotoxins and Related Fungal Metabolites. Chem. Rev. 109(9) 2009: 3903–3990; https://doi.org/10.1021/cr050001f.
9 TN-1119. Mycotoxins and Other Fungal/Bacterial Metabolites from Several Food Matrices by LC/MS/MS. Phenomenex: Torrance, CA, 2012; https://phenomenex.blob.core.windows.net/documents/c8b71f29-fc33-478b-ac0a-3950ee59d092.pdf.
Greg Allard is a principal scientist in process and analytical development, and corresponding author Sean Evans is senior vice president of manufacturing and development, and at writing Brian Pickering was associate director of quality control and microbiology — all at LFB USA in Framingham, MA; 1-508-494-8773; sean.evans@lfb-usa.com. Their former colleague Nick Masiello is now with LakePharma in Hopkinton, MA.