Cell-based advanced therapies are changing modern medicine dramatically. Immunotherapies such as chimeric antigen receptor (CAR) T-cell therapies are treating different forms of cancer. Gene therapies are reversing the course of inherited diseases, and tissue-engineered medical products are restoring, maintaining, and replacing damaged organs (1–4). The development of new advanced therapies is booming. As of January 2020, the US Food and Drug Administration (FDA) has reported more than 900 investigational new drug (IND) applications for cell and gene therapy products.
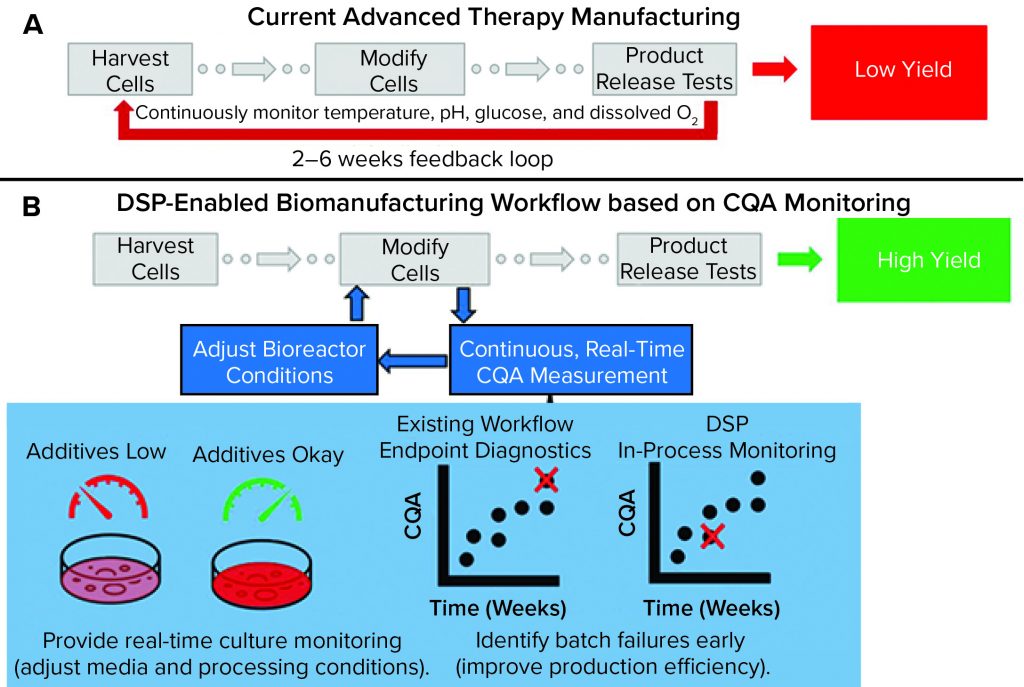
Figure 1: Comparing a representative state-of-the-art manufacturing workflow and one enabled by the dynamic sampling platform (DSP); (a) for cell therapy production, cells are harvested from a patient (autologous) or donor (allogeneic) before undergoing numerous steps, including those that “modify” cells to become medically potent. After a long production process, release assays are performed to determine whether a drug product can be delivered to a patient. Without real-time sensors to provide detailed information about process trajectory, processes often must be started over if they fail after running for many weeks. That situation increases cost and lowers yield. (b) The DSP workflow enables real-time sensing of critical quality attributes (CQAs) for improved process control and higher production yield. In the near term, CQA information can be used to predict early failure such that processes can be started over earlier, thereby improving the failure-to-treat ratio and ensuring that patients are treated as soon as possible.
However, such potentially transformative treatments have yet to reach broad patient populations despite having seemingly similar production workflows to widely adopted biologics. In the well-established biopharmaceutical industry, a medically potent biomolecule produced by a cell is the final product. But in the nascent cell therapy space, the entire cell is the final product, comprising thousands of different biochemicals. That difference makes characterization and control during product manufacturing far more challenging to ensure the safety, efficacy, and potency of cellular therapies.
Figure 1a shows a representative state-of-the-art cell therapy manufacturing workflow (e.g., for a CAR-T product). It includes harvesting cells from a patient or donor (e.g., leukapheresis), modifying (e.g., viral transduction) and expanding those cells to create a clinically potent and sufficiently sized therapeutic product, and releasing that therapy based on an endpoint assay (e.g., cytokine release test). Currently, few (if any) real-time analytics are available to serve as indicators of production trajectory. The result is a suboptimal, low-yield process design that relies on endpoint diagnostics to verify product quality. Such a process can create extremely long feedback loops with high production costs (>50% of CAR-T production cost attributed to materials and supplies), which are detrimental to patients who do not have the lifespan to wait for production to start over (5). In fact, the lack of process analytical tools (PATs) suitable to serve as inputs for feedback control is the main bottleneck to mass-market entry and contributes substantially to high patient costs (US$0.5–2.0 million per treatment) associated with the few FDA-approved regenerative medicine techniques (6).
Predicting whether a therapeutic product will be potent, efficacious, and safe is challenging for advanced therapy workflows because the critical quality attributes (CQAs) that indicate product quality remain relatively undefined. Some of the most reliable CQAs for predicting cell state (and thus product quality) are the secreted signaling and paracrine factors that cells release while developing in culture (7–9).
The dynamic sampling platform (DSP) is a PAT for real-time CQA biomolecule detection, discovery, and monitoring. It is being developed to fit into existing biomanufacturing workflows and provide detailed information about secreted CQA biomarkers. As Figure 1b shows, the DSP enables measurement of CQA content that benefits advanced therapy workflows by enabling feedback for improved process control (thus helping to increase yield). For instance, detailed readouts on secreted CQAs can be used to adjust and control media composition, ensuring that optimal growth conditions are met with minimal material waste. Correlating CQA content with final product trajectory should enable early detection of batch failure. The type of sensing provided by the DSP can improve process efficiency, decrease therapy costs, and reduce failure-to-treat ratios.
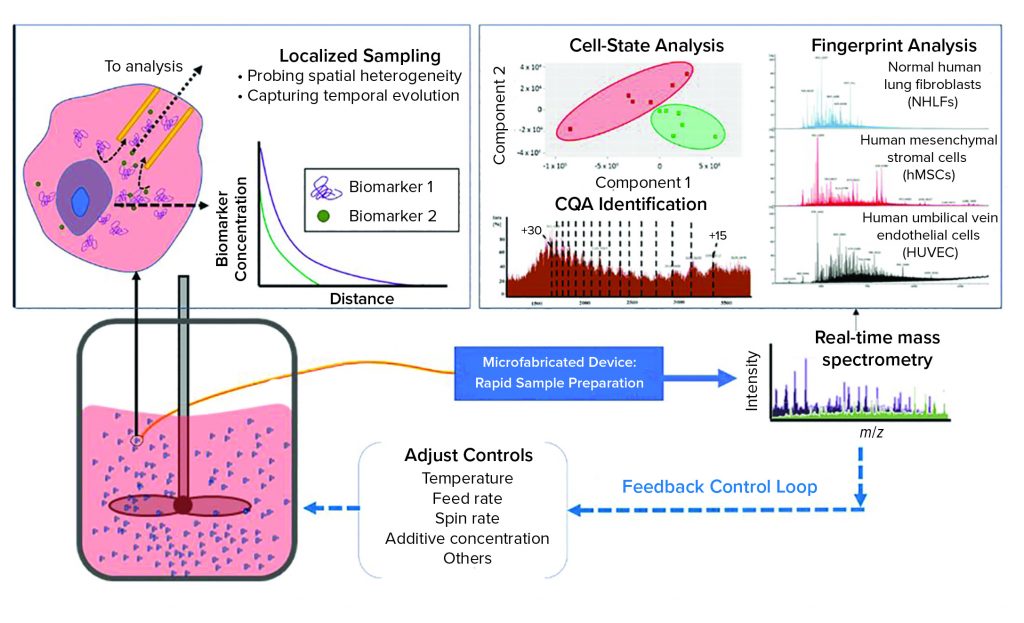
Figure 2: The dynamic sampling platform (DSP) enables direct sampling from a bioreactor with high spatial resolution to capture biomarkers that indicate product quality and process status. This sample, containing both biomolecules of interest and interfering species, is conditioned rapidly for analysis within a microfabricated device, facilitating small-volume operation with high throughput. The DSP technology can integrate with a range of sensing techniques, with electrospray ionization mass spectrometry (ESI-MS) being the most powerful because of its high sensitivity and label-free detection. The output can be used to predict cell states (e.g., proliferation, differentiation), to identify the biomarkers that serve as critical quality attributes (CQAs), and it can be used to provide fingerprint analysis of complex systems. The DSP is rapid (about 1 minute from sample to analytical output), so it can serve as in input for feedback control. (inset) The cell microenvironment has a high relative concentration of secreted biomarkers that correlate with cell state. These biomarkers are rapidly diluted away from the cell, making detection difficult for traditional process analytical technologies that capture only bulk media. DSP samples locally from the cell microenvironment to capture heterogeneities that can influence cell behavior. By sampling near the cells, DSP captures temporal changes in the secretome such that the output is a direct representation of real-time cell state.
DSP As an Enabling PAT for Cell Therapy Manufacturing
Widely adopted real-time PATs, including those that measure pH, dissolved oxygen (O2), glucose, and temperature sensors are necessary to maintain culture viability, but they do not provide information about the low abundance CQAs (e.g., secreted biomarkers) that more accurately predict cell state. Within the bioprocess industry, successful adoption of sensitive real-time PATs involving Raman spectroscopy has been effective for targeted monitoring of certain biomolecules relevant to large-molecule production. But such technologies lack the sensitivity to detect most low-concentration biomarkers continuously secreted by cells (10, 11). The most effective methods to characterize secreted CQA biomarkers are generally off-line. Such technologies include high-performance liquid chromatography–mass spectrometry (HPLC-MS) and enzymatic assays (e.g., enzyme-linked immunosorbent assay, ELISA). Off-line methods are limited in their utility as inputs for feedback control. Typically, such approaches for CQA characterization are used only to monitor bulk media, and the results represent a temporal and spatial average of secreted CQAs.
Bioreactors are highly heterogeneous environments in both space and time. That variability presents a significant challenge for sensors that seek to monitor CQAs within a bioreactor volume. Although many production-scale bioreactors are stirred for homogenization, the cell microenvironment remains highly dynamic. The inset image in Figure 2 illustrates how secreted biomarkers (CQAs) vary as a function of distance from cells. In close proximity to a cell, (within the microenvironment), secreted biomarkers are in high relative concentration compared with other media components that generally dominate analytical outputs based on bulk media samples.
The DSP leverages that local CQA enrichment by sampling from within the cells’ microenvironment to capture CQAs as they are secreted. That is beneficial for characterizing bioreactor cultures because sampling in a cell microenvironment enables probing of cell-to-cell heterogeneity, and the microenvironment provides an instantaneous picture of cell state. Traditional approaches that analyze only bulk media cannot identify when or where a particular CQA originated. Thus, they represent average measures of CQA content and bioreactor status. By contrast, cellular signaling (used to predict cell state accurately) occurs on the scale of single cells.
Other approaches to in-line bioreactor monitoring using MS monitoring have fallen short because they are spatially unresolved, lack consistency of analytical output, or present challenges with scaling up to process a large number of samples. The DSP is made through batch microfabrication. So the DSP manufacturing process is scalable for high-volume production, and it ensures that each device is identical for highly repeatable results. The microfabricated DSP has an active working volume of about 20 nL, which enables high-throughput analysis with a nearly one-minute response time.
The DSP samples directly from bioreactors with single-cell spatial resolution, enabling detailed probing of bioreactor heterogeneity. The method already has shown that it can integrate with bioreactors for cell-state monitoring throughout an entire growth cycle without introducing contamination (13). By using MS as a sensing method, the DSP can be used in a CQA-discovery mode to detect and identify biomarkers down to the attomolar range, addressing a key shortcoming of common real-time PATs
As Figure 2 shows, the DSP samples directly from a bioreactor with high spatial resolution to capture significant CQA heterogeneity within the volume. Small samples of extracted media are treated for analysis within the microfabricated DSP conditioning module, which is coupled directly to an outlet enabling real-time analysis by electrospray ionization mass spectrometry (ESI-MS) (12).
As a platform technology, the DSP can be coupled with the appropriate analytical techniques for given applications. In cellular manufacturing applications, CQAs include extremely low concentration (atto-nanomolar) biomarkers contained within media comprising inorganic salts and other biomolecules (e.g., serum additives) that are orders of magnitude higher in concentration (millimolar). A fundamental challenge for advanced therapy development is to enable both the monitoring of known CQAs and the discovery of CQAs that have not yet been identified.
An effective analytical output for advanced therapy manufacturing should be highly sensitive with a wide dynamic range while allowing for label-free biochemical detection. Thus, ESI-MS is an excellent sensing choice and has established use for characterizing biomolecules ranging in size from metabolites to large proteins.
For most biological analyses, ESI-MS is coupled with an off-line separation technique such as HPLC. Samples are conditioned to reduce spectral overlap caused by multicomponent mixtures and to remove compounds (e.g., inorganic salts) that suppress biochemical signatures and complicate identification. The DSP replaces such time-consuming sample preparations in unit-operation format with a flow-through microfabricated device for continuous sample treatment that enables rapid and highly sensitive detection of CQAs in situ in cell culture media (12, 13).
The DSP has been applied to different live cell cultures for real-time ESI-MS analysis. It has demonstrated that significant biochemical heterogeneities exist even within simplistic two-dimensional cell cultures such as six-well plates. Those experiments revealed that only with localized sampling (within one cell diameter of the membrane) can the DSP detect differences in secretome associated with phenotypically different cell types. The platform technology also has been applied to cells throughout their entire culture process (about 21 days) for continuous monitoring without contaminating cultures or affecting final cell states. Careful analysis of the results revealed that cells can be detected in states that are relevant to manufacturing, including proliferation, confluence, and differentiation (13).
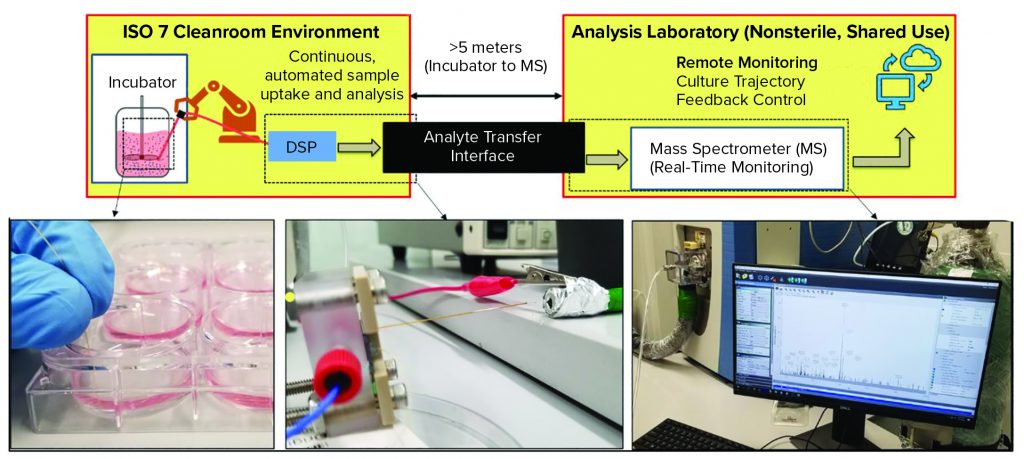
Figure 3: Enabling DSP for good manufacturing practice (GMP) compatibility is a key challenge for the technology. Advanced therapies, like all pharmaceutical products, must be produced in facilities that adhere to strict GMP guidelines, which govern nearly every aspect of manufacturing. Using DSP within GMP can be difficult because the technology should be near where cells are cultured (e.g., sterile ISO-7 environments) whereas the analytical output is in a separate, shared-use facility. Using an analyte transfer interface, DSP has been integrated with a GMP environment located at Georgia Institute of Technology. For a full, pilot-ready system, the DSP should be automated for sample uptake and analysis, and the output should enable remote monitoring of the culture trajectory and provide feedback control. (inset, left): Photo shows manual samples taken from a six-well plate. (inset, middle) The DSP, within a fluidic package for easy handling, infuses sample directly into the analyte transfer interface inlet within the GMP environment. (inset, right) The outlet of the analyte transfer interface couples to the mass spectrometer for real-time analysis.
Integration with CGMP and Continuous Monitoring
For commercial-scale relevance, the DSP must provide continuous monitoring of cells with minimal disruption of production workflows in a good manufacturing practice (GMP) environment. At commercial scales, cells are cultured inside sterile bioreactors typically housed within low–particle-count environments (e.g., ISO-7 or class 10,000 cleanrooms). Analytics most often are carried out in shared-use laboratory facilities that are separate from production processes.
The schematic in Figure 3 illustrates the layout of the GMP-like facility at the Georgia Institute of Technology. Inset images illustrate key features of the DSP-enabled PAT for GMP integration. After samples are taken from a cell culture (Figure 3 inset, left), they flow through the DSP’s microfabricated mass exchanger for sample conditioning and subsequent ESI of biomarkers for MS analysis through the long-distance analyte transfer interface (Figure 3 inset, middle). Ion transfer in gas phase enables fast delivery to the MS inlet for real-time data analysis located in the analytical suite (Figure 3 inset, right) (14).
The DSP has demonstrated key milestones for use in advanced-therapy production. The method addresses major challenges that nearly all PATs face by providing both spatial and temporal resolution in a minimally invasive way. For more complex bioreactors and continuous monitoring of commercial-scale systems, the DSP should be refined further and automated for positioning and sample extraction. Automation also reduces the number of operators required because an entire DSP system can be automated from uptake to analytical output continuously.
In addition to efforts toward automation, developing a greater understanding of how real-time DSP output correlates with a product’s therapeutic quality is underway. No single biomarker indicates the quality of an advanced therapy, and thus it is necessary to implement PATs that can simultaneously monitor multiple target biomarkers. Real-time “fingerprint” data gathered by the DSP represents a wide range of biomarkers simultaneously. To validate this output fingerprint data, it should be compared with standard assays in biomanufacturing workflows. Traditional assays (e.g., ELISA, immunoassays, and microarrays) enable multiplexed analysis in which multiple biomarkers are assessed simultaneously. So it is advantageous for the DSP system to operate in multiplex format for monitoring advanced-therapy workflows. When coupled to ESI-MS, the DSP is a highly capable multiplexing tool because the output mass spectra provide information on all biomarkers present in solution, with no specific labeling required.
Practical concerns such as limited dynamic range, matrix and ionization effects, and spectral overlap make unambiguous detection of every biomarker challenging. So time-consuming sample preparation (e.g., tryptic digestion, HPLC) typically is performed for MS workflows. The DSP does not have dynamic range limitations because it captures secreted, quality-indicating biomarkers at their source, where they are in highest relative abundance. To mitigate matrix interference, analyte-dependent ionization efficiency effects, and/or spectral overlap, the DSP sample treatment protocol can be tuned to improve limits of detection and selective identification of a broad range of analytes (12, 13). Alternatively, the DSP can operate in “multiple singleplex” mode for targeted applications such that a specific sample treatment protocol is optimized to detect target CQA biomarkers (including quantification). This mode of operation is similar to other targeted assays (e.g., antibody-based sensing) that are optimized for highly specific and sensitive detection of certain biomarkers (15).
It is important to develop the DSP such that the analysis method is optimized for the application. Feedback control requires advanced understanding of how inputs that control cell culture (e.g., temperature, pH, and additives) affect final product quality and intermediate biochemical states. Efforts are underway to build that knowledge, and that will enable real-time DSP outputs to serve as effective and accurate measures of culture trajectory as well as inputs for feedback control. With automation and remote monitoring capabilities, the DSP can enable scale-up and scale-out of advanced therapy production.
Improved Biomarker Monitoring
For advanced therapies to reach a broad patient population, their manufacturing must be optimized in terms of cost, yield, and success rate. Unfortunately, many promising therapies fail to achieve FDA approval because their sponsors lack understanding of the chemistry, manufacturing, and controls (CMC) standards that govern production. Existing PATs for biomanufacturing are poor at predicting safety, efficacy, potency, and overall quality of complex advanced therapies. The DSP addresses those challenges as an integrated PAT system designed for use in bioprocess workflows. It can detect cells in states that are relevant to manufacturing by using ESI-MS sensing of CQAs that are captured at their source by probing a cell microenvironment.
As a discovery tool, the DSP enables process understanding for more robust CMC standards by identifying CQAs that correlate with cell therapeutic quality. By providing a minute-scale temporal response, the DSP also serves as a valuable real-time PAT for process monitoring applications. The technology already has been integrated with GMP workflows, which significantly reduces the amount of effort required for integration into commercial-scale workflows. Our ultimate goal is to fill the void of effective PATs for bioprocessing such that current and future production systems for advanced therapies can be scaled up and scaled out for more available and affordable treatments.
Acknowledgments
We acknowledge Austin Culberson and Peter A. Kottke for their support in developing the DSP and its auxiliary systems. We thank Krishnendu Roy, Carolyn Yeago, and the rest of the Marcus Center for Therapeutic Cell Characterization and Manufacturing (MC3M) staff members, including Hazel Stevens, Annie Bowles, Linda Kippner, Nina Li, and Paramita Chatterjee for assisting with MS acquisition and installation, establishing DSP GMP compliance, and providing guidance for advanced therapy processes. The work described herein is supported by NSF Center for Cell Manufacturing Technologies (CMaT) Award 1648035, Marcus Center for Therapeutic Cell Characterization and Manufacturing Collaboration Grant in Cell Manufacturing, the Georgia Tech Foundation, and the Georgia Research Alliance. Partial support was provided by Grant Number RO1GM112662 from the National Institute of General Medical Science (NIGMS), a component of the National Institutes of Health (NIH). These contents are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS or NIH. Device microfabrication was performed in part at the Georgia Tech Institute for Electronics and Nanotechnology, a member of the National Nanotechnology Coordinated Infrastructure, which is supported by the National Science Foundation (Grant ECCS-1542174).
Conflict of Interest
Mason Chilmonczyk and Andrei Fedorov are inventors of the technology being studied. The purpose of this project is to explore its commercialization. The terms of this arrangement have been reviewed and approved by Georgia Institute of Technology in accordance with its conflict of interest policies.
References
1 Iyer NR, Thomas SW, Sakiyama-Elbert SE. Stem Cells for Spinal Cord Injury: Strategies to Inform Differentiation and Transplantation. Biotechnol. Bioeng. 114(2) 2017: 245–259; https://doi.org/10.1002/bit.26074.
2 Kaiser AD, et al. Towards a Commercial Process for the Manufacture of Genetically Modified T Cells for Therapy. Cancer Gene Ther. 22, 2015: 72–78; https://doi.org/10.1038/cgt.2014.78.
3 Oettgen P. Cardiac Stem Cell Therapy. Circulation 114(4) 2006: 353; https://doi.org/10.1161/CIRCULATIONAHA.106.639385.
4 Poulos J. The Limited Application of Stem Cells in Medicine: A Review. Stem Cell Res. Ther. 9(1) 2018: 1; https://doi.org/10.1186/s13287-017-0735-7.
5 Harrison RP, et al. Chimeric Antigen Receptor T Cell Therapy Manufacturing: Modelling the Effect of Offshore Production on Aggregate Cost of Goods. Cytother. 21(2) 2019: 224–233; https://doi.org/10.1016/j.jcyt.2019.01.003.
6 Aijaz A, et al. Biomanufacturing for Clinically Advanced Cell Therapies. Nature Biomed. Eng. 2(6) 2018: 362–376; https://doi.org/10.1038/s41551-018-0246-6.
7 Mucida D, et al. Reciprocal TH17 and Regulatory T Cell Differentiation Mediated By Retinoic Acid. Science 317(5835) 2007: 256–260; https://doi.org/10.1126/science.1145697.
8 Agarwal S, Rao A. Modulation of Chromatin Structure Regulates Cytokine Gene Expression During T Cell Differentiation. Immunity 9(6) 1998: 765–775; https://doi.org/10.1016/S1074-7613(00)80642-1.
9 Albrecht S, et al. Proteomics in Biomanufacturing Control: Protein Dynamics of CHO–K1 Cells and Conditioned Media During Apoptosis and Necrosis. Biotechnol. Bioeng. 115(6) 2018: 1509–1520; https://doi.org/10.1002/bit.26563.
10 Abu-Absi NR, et al. Real-Time Monitoring of Multiple Parameters in Mammalian Cell Culture Bioreactors Using an In-Line Raman Spectroscopy Probe. Biotechnol. Bioeng. 108(5) 2011: 1215–1221; https://doi.org/10.1002/bit.23023.
11 Thiagarajan G, et al. Use of Raman and Raman Optical Activity for the Structural Characterization of a Therapeutic Monoclonal Antibody Formulation Subjected to Heat Stress. J. Raman Spec. 46(6) 2015: p. 531–536; https://doi.org/10.1002/jrs.4679.
12 Chilmonczyk MA, et al. Dynamic Mass Spectrometry Probe for Electrospray Ionization Mass Spectrometry Monitoring of Bioreactors for Therapeutic Cell Manufacturing. Biotechnol. Bioeng. 116(1) 2019: 121–131; http://doi.org/10.1002/bit.26832.
13 Chilmonczyk MA, et al. Localized Sampling Enables Monitoring of Cell State via Inline Electrospray Ionization Mass Spectrometry. Biotechnol J. 24 September 2020; https://doi.org/10.1002/biot.202000277.
14 Garimella S, et al. Gas-Flow–Assisted Ion Transfer for Mass Spectrometry. J. Mass Spec. 47(2) 2012: 201–207; https://doi.org/10.1002/jms.2955.
15 Christians U, et al. Mass Spectrometry-Based Multiplexing for the Analysis of Biomarkers in Drug Development and Clinical Diagnostics: How Much Is Too Much? Microchem. 105, 2012: 32–38; https://doi.org/10.1016/j.microc.2012.02.011.
Mason A. Chilmonczyk is a postdoctoral researcher at the George W. Woodruff School of Mechanical Engineering, Georgia Institute of Technology and at the NSF ERC Center for Cell Manufacturing Technologies (CMaT), Parker H. Petit Institute for Bioengineering and Biosciences, Georgia Institute of Technology. Corresponding author Andrei G. Fedorov is a professor and Rae S. and Frank H. Neely chair at the George W. Woodruff School of Mechanical Engineering,and at CMaT; Georgia Institute of Technology, 771 Ferst Dr., Atlanta 30332-0405, GA; andrei.fedorov@me.gatech.edu.
