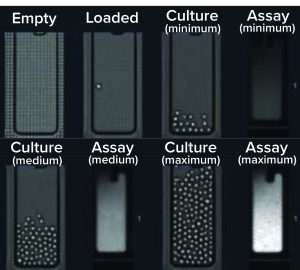
The Opto CLD workflow provides a complete visual record from single-cell cloning into NanoPen chambers through recovery of selected clones into well plates. (https://www.berkeleylights.com/workflows/cell-line-development)
On 1 December 2020, BPI presented an “Ask the Expert” webinar with Tanner Nevill (vice president of program management at Berkeley Lights). Monoclonality assurance is a central regulatory requirement for all cell lines producing biologic therapies. Well-plate imaging is the “gold standard” for confirming that a cell line originates from a single cell, but it is labor intensive and prone to errors in the form of “ghost cells” and debris that look like cells. Using the Berkeley Lights Beacon system, cells are cloned, cultured, and assayed over multiple days directly on a microfluidic chip. The system provides a complete visual record from initial single-cell cloning through recovery of selected clones into well plates. Cell lines are recovered with >99% monoclonality assurance in under a week, removing the need for multiple lengthy rounds of cloning.
Nevill’s Presentation
The Opto CLD workflow provides visual verification of clonality at every stage through multiple days of ownership culture. It begins with automated loading of transfected pools from well plates onto nanofluidic OptoSelect chips in the Beacon system. Each chip contains thousands of NanoPen chambers <2 nL in volume, which enables efficient growth and detection of secreted antibody molecules.
OptoSelect technology manipulates cells with light to identify them automatically and move them in parallel into individual NanoPen chambers. Thousands of cells can be cloned on a single chip in under three hours, and the system can handle up to four chips at a time.
After cloning, cells are perfusion cultured on the chip. Media diffuses into each NanoPen chamber while waste products diffuse out and are swept away. Diverse growth rates and phenotypes can be observed from a single transfected pool, and clones are imaged throughout the culture to provide visual confirmation of clonality.
To select top clones, the Opto CLD workflow uses a fluorescent small molecule that binds to secreted antibodies in the pens. After unbound reagent is flushed out, pens with the most secreted antibodies will have the brightest signals. This assay is quantitative over a wide range of secretion levels and can be repeated over multiple days. With automated cell counts, you can monitor cell growth rates and calculate cell-specific productivity. Filters can be used in the accompanying software package to select and rank clones based on productivity, growth rate, and other phenotypes.
In the last step, desired clones are recovered from the NanoPen chambers using the same light-based manipulation and deposited into individual wells of a 96-well plate. The recovery process incorporates in-line controls and imaging verification steps to ensure clonality is maintained.
Berkeley Lights has tested clonal recovery. Only two wells out of nearly 1,300 were found to contain cells. That translates to >99% clonality assurance with very high outgrowth.
Questions and Answers
What have regulatory agencies said about Opto clonality assurance? Currently, multiple customers have molecules in clinical trials with cell lines selected using the Beacon system. The US Food and Drug Administration (FDA) has given favorable feedback on monoclonality assurance with the Beacon system, and >99% monoclonality value has been validated in a peer-reviewed publication from Amgen.
Does Berkeley Lights have experience with suspension cell lines? Most groups using the Berkeley Lights platform are working with suspension cells, but a few work with adherent cells. That requires a different surface chemistry. Coating can be applied to the insides of the pens but not to the main channel so that adherent cells will grow exclusively inside the NanoPen chambers.
Is there risk of cells moving from chamber to chamber? We have exhaustive video records of cells cultured over time, with evidence showing no travel from pen to pen during that process. Well-to-well cross contamination is <1%, with clonality maintained over 99% of the time.
Can Berkeley Lights systems be used for non-CHO cell lines? We’ve worked with many mammalian cells, and now we are working with yeast and bacteria. Our platforms are agnostic to cell and media type.
