Photo 1: Twin-column equipment Contichrom CUBE Combined (BENCHTOP SCALE, LEFT) and EcoPrime Twin (PILOT SCALE, RIGHT)
Periodic countercurrent (PCC) processes increasingly are being evaluated as alternatives to single-column batch capture processes. Some of the advantages of PCC processes over single-column processes include shortening of processing time and/or reduction of required resin volume through increased productivity; reduction in resin costs through improved resin capacity use; and reduction in buffer consumption through increased column loading. Those advantages, however, come with increased equipment complexity and hardware costs. PCC processes and systems with two to up 16 columns of the same type have been proposed. As the number of columns increases, a process becomes more complex, potentially adding pumps, detectors, valves, and plumbing. The PCC process investigated in this study was a CaptureSMB process: a PCC process using two columns and modulated feed flow rates. All PCC processes are based on the concept of loading the first affinity column beyond its dynamic breakthrough capacity and capturing what breaks through in a second column of the same type. So the first column can be loaded to values of 50â80% breakthrough, whereas in batch chromatography, loading is typically performed to values below 1% breakthrough to prevent product loss (1). Based on the adsorption kinetics and mass-transfer properties of commercially available protein A resins, interconnecting more than two columns in the load zone provided little improvement in performance (2, 3).
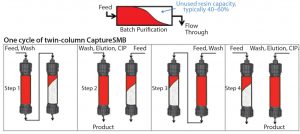
Figure 1: Schematics of batch purification (TOP) and one cycle of the CaptureSMB process (BOTTOM); CIP = clean in place
Baur et al. compared the performance of CaptureSMB, three- and four-column PCC processes based on numerical optimization (4). Results showed that a high capacity use (>95%) was achieved across all formats, which is expected because all formats adopt the above-mentioned loading of two columns in series. The authors showed that among the three formats tested, CaptureSMB offers productivity advantages, especially with low and high expression titers. That is because the interconnected loading phase of that process can be adjusted in duration independently of other process steps as titer increases or decreases. Figure 1 outlines the principle of the CaptureSMB process schematically based on one process cycle.
In the first step, the two affinity columns are interconnected. The first column is loaded beyond its dynamic breakthrough capacity, followed by a buffer wash to transfer unbound product from the first column to the second column.
In the second step, the two affinity columns are operated in parallel as single columns. While the first (overloaded) column is washed, eluted, cleaned, and reequilibrated, the second column continues loading until it reaches 1% breakthrough. The loading flow rate of the second column can be reduced to synchronize with recovery and regeneration of the first column.
In the third step, the second and first columns are interconnected and loaded in series analogous to the first step.
In the fourth step, the second column is washed, eluted, cleaned, and reequilibrated while loading continues on the first column.
After one cycle is complete, the process can be continued over multiple cycles. An interconnected phase with additional load (which is calculated based on knowledge of the product breakthrough curve) starts up the cyclical steady-state process, taking into account that the columns are initially devoid of product. When the process is shut down, the first column is washed, eluted, cleaned, and reequilibrated while the second remains inactive (because it was cleaned and reequilibrated in the previous step).
Previously conducted studies have focused on evaluating PCC processes at laboratory and benchtop scales (5, 6). Scalable multicolumn processes are used in pharmaceutical applications mainly for separation of small-molecule enantiomers (using simulated moving bed, SMB, chromatography) (7, 8). However, the greater complexity of requirements for biologics purification has raised questions about SMB scalability. Biologics are more heterogeneous than small-molecule drugs in active pharmaceutical ingredient (API) composition, generally are more susceptible to degradation and aggregation, and can contain complex product- and process-related impurities in the starting material.
Often, several orthogonal steps, each consisting of a distinct separation mechanism, are needed to purify a product to target specifications. Of those steps, protein A capture is the primary workhorse for removing most process-related impurities in monoclonal antibody purification. However, that affinity-chromatography step has become the most expensive step during downstream processing because of the high cost of protein A resin as well as limited options for operating in batch mode. PCC processes such as CaptureSMB overcome that challenge by enabling high-capacity use at rapid flow rates. The data presented here demonstrate a successful scale-up of the process from benchtop to pilot and production scales.
Materials and MethodsÂ
For benchtop-scale experiments, a Contichrom CUBE Combined 30 system (ChromaCon, ZĂŒrich, Switzerland) was used. The system can run single-column batch processes and twin-column processes such as CaptureSMB, twin-column polishing (multicolumn solvent gradient purification, MCSGP), and integrated two-step batch chromatography. Pilot-scale runs used an EcoPrime Twin 100 system (LEWA-Nikkiso America, Inc.). The EcoPrime Twin system has a sanitary design and can manufacture in compliance with current good manufacturing practice (CGMP) standards. This system also can run chromatography in single-column batch mode and in two-column interconnected continuous mode (e.g., AEX-CEX). Both systems have UV, conductivity, and pH detection capabilities (Photo 1, first page).
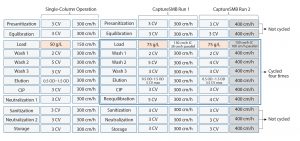
Based on breakthrough curve data generated separately, the CaptureSMB process design uses the wizard included in the Contichrom CUBE operating software (ChromIQ). Based on the input data, the wizard generated the CaptureSMB methods to be run including all valve settings, loading, washing and regeneration flow rates, and corresponding startup and shutdown methods. The small-scale experiments had two columns packed with MabSelect SuRe LX resin (GE Healthcare) with dimensions of 1.0-cm i.d. Ă 10.0-cm bed height (CV = 7.85 mL for each column). The scale-up experiments used two 10.0-cm i.d. Ă 10.0-cm bed-height columns (CV = 785 mL for each column) packed with the same resin, representing a scale-up factor of 100Ă in terms of column volume. The batch reference experiment used a column of 24.5-cm bed height and 30-cm i.d. (CV = 17,300Â mL) packed with MabSelect SuRe LX resin. The starting material for each run was a clarified cell culture harvest containing IgG4 with a titer of 5 g/L. Figure 2 shows the single column reference process and the CaptureSMB processes.
Column loading for the single-column batch chromatography was limited to 50 g/L based on 1% breakthrough data generated before the run. Flow rates for load and remaining steps were optimized based on maximizing the loading capacity and improving productivity, respectively. Two CaptureSMB runs were performed, a âstandardâ and an âoptimizedâ run (Run 1 and Run 2, respectively). In the standard run, all scaling parameters were maintained constant between the batch-mode chromatography and the CaptureSMB chromatography except for the higher loading capacity and operation in cyclical mode. Loading capacity was optimized to 75Â g/L based on 60% breakthrough with an additional safety margin to prevent any product losses.
Considering the length of time spent in eluting and regenerating the columns, operating flow rates in parallel mode (Steps 2 and 4 in Figure 1) were increased from 300 cm/h to 400 cm/h in the optimized run. Consequently the loading flow rate was increased to 100 cm/h rather than 30 cm/h in standard operation. Such higher flow rates are possible in CaptureSMB because of lower bed heights and, consequently, lower backpressure across the column. As a result, the cycle durations were reduced from 230 to 170 minutes (four cycles in total) for the optimized run using a total resin volume of 1.57 L. Both runs included start-up and shutdown methods. By comparison, batch chromatography lasted 260 minutes for the operation using 17.3-L total resin volume. Product elution fractions, collected separately, were analyzed for titer, host cell protein (HCP), DNA, leached protein A ligand (rProA), and aggregate content.
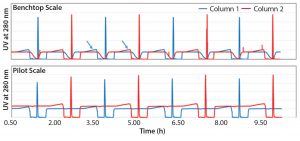
Figure 3: UV profiles (280 nm) at benchtop and pilot scale (Run 2), with arrows indicating the breakthrough of unbound monoclonal antibody
Results
Continuous capture chromatography experiments were run at benchtop and pilot-plant scales using initial and optimized process parameters. UV profiles were comparable for both runs (Figure 3 ). At both scales, repeating patterns of alternating product elution (sharp peaks in the chromatograms) were recorded by the UV detectors behind each column. Likewise, the UV detectors recorded the breakthrough curves during the interconnected loading phases. Because of the high resin use by the upstream column, during the interconnected loading phase not only the nonbinding impurities, but also the MAb breaks through from the upstream column. Unbound MAb, which can be identified by a rising UV signal during the last part of the breakthrough curve (see arrows), then gets captured on the downstream column.
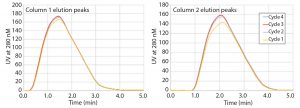
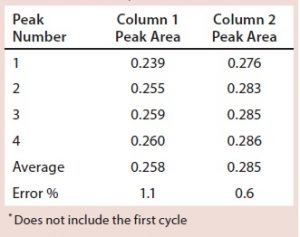
Figure 4 shows the elution peaks of pilot Run 1 overlaid separately for each column. The profiles are almost identical; only the first peaks have slightly smaller areas due to the approximation of the preload for the start-up loading phase.
Table 1 lists peak areas collected during elution on each column. A small variability in the UV detector calibration shows area values of the column 1 elution to be smaller, but the values are reproducible for each column with an error of 1.1% or less.
At pilot scale, Run 1 of CaptureSMB processed about 450 g of product from the clarified harvest in 19 hours using 1.57 L of protein A resin. Upon improvements in processing speed, a similar amount of product was purified in 14 hours using 1.57 L of protein A resin during Run 2 (Figure 2). By comparison, the batch chromatography captured a similar load in 3.5 hours using 17.3 L of protein A resin, but at a lower binding capacity. This corresponds to approximately 2.5-fold higher productivity (grams of MAb produced per hour per liter of resin) in the optimized CaptureSMB run compared with the batch capture chromatography. All benchtop and pilot-scale runs were evaluated with respect to productivity, capacity use (dynamic binding capacity/static binding capacity), and buffer consumption (liters of buffer used per grams of MAb produced) (Figure 5). Yields of all runs were â„94%.
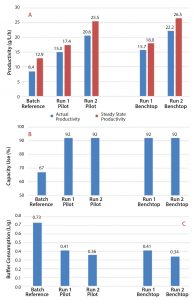
Figure 5: Productivity (A), capacity use (B), and buffer consumption (C) for pilot and benchtop scale CaptureSMB operation, along with batch reference performance calculations
Figure 5A displays the productivities of the batch and continuous runs. The latter are also shown for the different scales and process conditions (Runs 1 and 2). The graphs also show actual productivities (blue bars) in comparison with the steady-state productivities (orange bars). The reported actual productivity values include the startup and shutdown of the operation rather than the runâs steady-state phase. During steady state, product concentration and quality remain constant from cycle to cycle.
Comparing the steady-state values showed a 1.8 fold improvement in productivity from a traditional batch process in the initial continuous process (Run 1). Further process optimization (Run 2) improved productivity by 2.5-fold. These improvements can be seen at both scales. Therefore, initial feasibility studies on the benchtop are verified at the pilot scale. We saw similar effects in resin use at both scales, in which capacity use increased significantly from 67% to 92% for continuous capture (Figure 5B). The buffer consumption (Figure 5C) was reduced by 50% compared with that for batch chromatography. Again, similar results were obtained for both scales during continuous capture.
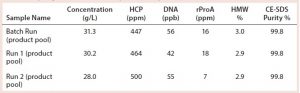
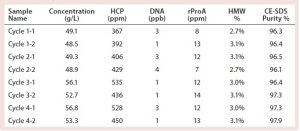
During all runs, the collected product pools were analyzed in terms of product concentration, HCP, DNA, rProA, and high molecular weight (HMW) species content. These critical quality attributes are comparable among batch and pilot scale CaptureSMB processes (Table 2). All results were well within the assay variability of the analytical methods.
Purity of elution peak samples collected during CaptureSMB Run 2 were further analyzed using in-line sampling valves on the EcoPrime Twin system. Good consistency in the impurity values was seen between elutions during the CaptureSMB operation, showing no signs of systematic deviations in performance quality (Table 3). The table shows results for each column (two elutions took place in each cycle), which explains some oscillations between values in every other sample. For this process, cyclic steady state is not attained until cycle 2, although the lower product concentrations at the beginning of the run could be attributed to the manual sampling procedure. Consistency in sampling can be attained by introducing automated sampling and PATs associated with it.
Discussion
A twin-column CaptureSMB process was successfully scaled up from laboratory to pilot scale by 100Ă for capture of a MAb from clarified cell culture harvest, with comparable process performance and quality attributes.
The process captured MAb from harvest material with 5-g/L titer, showing that the process can be used in conjunction with fed-batch upstream and not only with a continuous cell culture process (e.g., perfusion). This is an important aspect for clinical MAb manufacturing, in which rapid implementation of a continuous upstream step can depend on specific constraints in the manufacturing suite.
If the cadence for processing the cell culture harvest is maintained the same between a batch process and CaptureSMB process, a theoretical clinical batch of 20 kg (2,000-L bioreactor with a 10-g/L titer) could be processed in ~36 hours using a CaptureSMB process with about 25 L of resin distributed among two prepacked columns of 40-cm i.d. and 10-cm bed height. Conversely, using a single-column batch process would require a 100-L column with an 80-cm i.d. (20-cm bed height) operating for four cycles, taking about 36 hours, and consuming twice the amount of buffer. This example shows how the increased productivity of CaptureSMB supports use of smaller columns, potentially leading to significant resin savings.
Further, protein A resin often is left unused after a four- or five-lot campaign (16â20 cycles) in a clinical manufacturing setting until a repeat campaign occurs â which is typically not frequent. For batch purification, that results in severe underuse of the overall resin lifecycle and its available capacity. Alternatively, a CaptureSMB approach operates smaller columns over more cycles (60â80 cycles) per clinical manufacturing campaign. That again translates into major resin savings and effective use of resin capacity.
A Promising Alternative
In this scale-up study, buffer savings were around 50% using a PCC strategy. In the 20-kg MAb clinical trial scenario, that would correspond to savings of about 7,400 L. The question of process validation often is raised in conjunction with PCC processes. Without a doubt, the process validation effort increases proportionately with the number of columns. However, a preliminary risk-based validation concept design indicates that the effort would be manageable for a CaptureSMB operation (by contrast with column configurations using more columns) because much of the process parameters used in batch processing are directly scalable onto a CaptureSMB process. Further work is needed to qualify this operation fully as a replacement for traditional batch processing including resin lifetime cycling studies. The UV-based dynamic process control âAutomAbâ has been operated at PD scale and needs also to be validated on larger scale. Virus clearance and carryover studies have been carried out successfully for the CaptureSMB process.
Acknowledgments
The authors thank Srujana Govindarajulu, Danielle Belluscio, Kevin Potter, and Nicholas DiGioia for their help with pilot-scale operation and the Devens Process Development Analytics team for providing analytical support. Discussions with Professor Massimo Morbidelli and Dr. Daniel Baur from ETH ZĂŒrich (Swiss Federal Institute of Technology, ZĂŒrich) also are greatly appreciated.
References
1 Mahajan V, George A, Wolk B. Improving Affinity Chromatography Resin Efficiency Using Semi-Continuous Chromatography. J. Chromatogr. A 1227 (March 2012): 154â162; doi:10.1016/j.chroma.2011.12.106.
2 Baur D, et al. Optimal Model-Based Design of the Twin-Column Capture SMB Process Improves Capacity Utilization and Productivity in Protein A Affinity Capture. Biotechnol. J. 11(1) 2016: 135â145; doi:10.1002/biot.201500223.
3 Gjoka X, et al. A Straightforward Methodology for Designing Continuous Monoclonal Antibody Capture Multi-Column Chromatography Processes. J. Chromatogr. A 1416 (October 2015): 38â46; doi:10.1016/j.chroma.2015.09.005.
4 Baur D, et al. Comparison of Batch and Continuous Multi-Column Protein A Capture Processes By Optimal Design. Biotechnol J. 11(7) 2016: 920â931; doi:10.1016/j.chroma.2015.09.005.
5 Godawat R, et al. Periodic Counter-Current Chromatography: Design and Operational Considerations for Integrated and Continuous Purification of Proteins. Biotechnol. J. 7(12) 2012: 1496â1508; doi:10.1002/biot.201200068.
6 Warikoo V, et al. Integrated Continuous Production of Recombinant Therapeutic Proteins. Biotechnol Bioeng. 109(12) 2012: 3018â3029.
7 Miller L, et al. Chromatographic Resolution of the Enantiomers of a Pharmaceutical Intermediate from the Milligram to the Kilogram Scale. J. Chromatogr. A 849(2) 1999: 309â317.
8 Juza M, Mazzotti M, Morbidelli M. Simulated Moving-Bed Chromatography and Its Application to Chirotechnology. Trends Biotechnol. 18(3) 2000: 108â116.
James Angelo and John Pagano are scientists, corresponding author Srinivas Chollangi is a senior scientist, Xuankuo Xu is a principal scientist, Sanchayita Ghose is a director , and Zheng Jian Li is the executive director at Bristol-Myers Squibb, Inc., 38 Jackson Rd, Devens, MA 01434, USA. Thomas MĂŒller-SpĂ€th is COO at ChromaCon AG, Technoparkstr. 1, CH-8005 ZĂŒrich, Switzerland; and Kathleen Mihlbachler is global director of separations development at LEWA-Nikkiso America, Inc. Bioprocess Group, 8 Charlestown Street, Devens, MA 01434, USA.
This article was published as a preprint on BPIâs website in February 2018.