A large proportion of the therapeutic biotechnology products already in the market or under development are glycoproteins. Therapeutic glycoproteins are produced as recombinant products in cell culture systems, which raises the importance of understanding the biosynthetic events described in the previous installments of this three-part article. Lack of control in a bioprocess could easily change glycosylation patterns by distorting the activities taking place in the Golgi apparatus.
Disruption of the delicate balance among substrate availability, optimum pH for specific activities, glycosyltransferase and glycosidase location and availability, and so on can lead to products with different carbohydrate structures. Such parameters are different for different cell lines, and they can change with cell age and density of a culture as well as with the CO2, pressure, and the concentration of critical nutrients such as ammonium or metabolites such as butyrate. Changes in glycan structures can cause differences in protein stability and solubility, half-life in circulation, and possibly biological activity, folding, and aggregation.
WWW.PHOTODISCCOM
Modern technical advances allow the control of critical parameters to ensure that glycosylation takes place as expected to provide reproducible carbohydrate structures and in turn reproducible glycoprotein structures. In fact, it is possible to use this monitoring of glycosylation patterns as a means to establish process reproducibility and comparability between glycoproteins expressed by different systems and/or purified using different strategies.
PRODUCT FOCUS: GLYCOPROTEINSS
PROCESS FOCUS: ANALYTICAL
WHO SHOULD READ: QA/QC, PRODUCT DEVELOPMENT, PROCESS DEVELOPMENT, AND ANALYTICAL PERSONNEL
KEYWORDS: GLYCANS, CARBOHYDRATES, OLIGOSACCHARIDES, PAT, COMPARABILITY
LEVEL: INTERMEDIATE
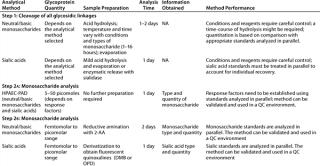
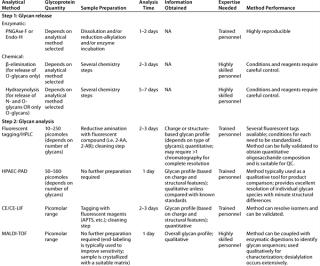
Evaluation of protein glycosylation implies first defining protein glycoforms, then individual glycan structures and their relative ratios, followed by site-specific glycosylation. Table 1 summarizes the most standardized tools for such purposes at the present time.
Table 1: Commonly used methods for the analysis of glycosylation in therapeutic glycoproteins by monosaccharide composition (all requiring the expertise of trained personnel)
Table 2: Commonly used methods for analysis of glycosylation in therapeutic glycoproteins by oligosaccharide mapping
Table 3: Commonly used methods for the analysis of glycosylation in therapeutic glycoproteins (site-specific glycosylation and glycoforms analysis)
When restricting the scope of carbohydrate analysis to a particular subclass of glycoconjugates, we can apply some general analytical strategies. Here I focus on the analysis of protein glycosylation. General principles for analysis of other glycoconjugate subclasses can be found elsewhere (8,9,10,11,12,13).
Unlike protein amino acid sequences, which can be derived theoretically from DNA sequences for recombinant proteins, there is no biochemical mechanism for predicting the complete theoretical carbohydrate composition of a glycoprotein. Therefore, it is impossible to orthogonally confirm the accuracy of carbohydrate measurements by comparison with theoretical values. And previously published literature values for well-known glycoproteins may not be as accurate as hoped due to the limits of technologies used at the time or quality practices at the research laboratories doing the work (e.g., use of qualified instruments, system suitability measures, validated methods, and diversity of test samples).
Monosaccharide Composition Analysis: Typically, glycoproteins produce diffuse, broad bands when separated in electrophoretic gels because of glycosylation heterogeneity. Treatment of glycoproteins with glycolytic enzymes, and electrophoresis followed by carbohydrate staining techniques or lectin blotting provides qualitative information about their glycosylation. These techniques are routine tools in protein laboratories, and they are useful at the research level and for developing purification strategies. Kits for lectin dot-blotting and lectin-based arrays for glycan analysis are also commercially available.
Measuring the precise molar ratio of monosaccharides, however, requires instrumentation and expertise. Monosaccharide composition analysis involves the following four steps: cleavage of all glycosidic linkages; fractionation
of the resulting monosaccharides; detection of each monosaccharide; and quantitation of the signals obtained.
A range of analytical methods have been developed over the years involving different cleaving strategies (hydrolysis with acid and methanolysis), analysis of native and derivatized monosaccharides (substituting polar groups producing volatile compounds or introducing fluorescent tags by reductive amination), and different separation principles (HPAEC, RP-HPLC, CE, and GLC) and detectors (PAD, fluorescence, FID) (11, 15, 16) To date, it has been difficult to systematize or standardize all these different methods even among expert analytical laboratories (17).
One reason such analyses are difficult to standardize is linked to the complexity of carbohydrate structures described in previous installments of this article. All monosaccharides are linked to each other through a glycosidic linkage. Cleavage of such linkages involving hexoses requires more vigorous conditions than cleavage of fucose or sialic acid linkages. The glycosidic linkage of glycosyluronic acids is particularly resistant to acid hydrolysis and requires special conditions (18, 19). Once monosaccharides are free in solution, their rate of decomposition also differs, creating a need for balancing the achievement of release while preventing complete destruction. So there is an inherent need for careful standardization of procedures using appropriate standards.
In recent years, two methods have emerged to be generally accepted as reliable:
-
analysis of native monosaccharides obtained by acid hydrolysis using a high-performance anion-exchange chromatography eluted with high-pH mobile phases and monitored by pulsed amperometric detection (HPAEC-PAD) (20)
-
analysis of derivatized monosaccharides obtained by acid hydrolysis followed by the introduction of a fluorescent tag (2-AA is best suited because it requires no re-N-acetylation of hexosamines after acid hydrolysis whereas other fluorescent tags do) through reductive amination using reverse-phase HPLC with online fluorescence detection (21).
Both approaches require systematic analysis of appropriate monosaccharide standards treated in parallel to the samples. Such techniques require properly set-up (IQ, OQ) and maintained (PQ) equipment, proficient analytical chemists, and well-written procedures for analyses to be performed. For accurate determination of monosaccharide content, they also require concurrent analysis of individual monosaccharides that are of known concentration. A wide range of monosaccharide standards are commercially available for incorporation into your analytical procedure of choice. When carefully executed, both approaches above can be validated for a specific glycoprotein to ensure method accuracy, reproducibility, intermediate precision, robustness, and ruggedness. Accurate, reliable method performance is necessary for product specifications to be established and support method implementation in QC environments.
Information obtained from either of those two methods represents only an overall ratio and content of monosaccharides in a given product. However, information about the consistency of antennary structures or distribution of species among glycosylation sites will be lost during the hydrolysis step needed to release monosaccharides for analysis. So this type of analysis — although quantitative and validatable — is often overpowered by oligosaccharide analysis strategies.
Another type of monosaccharide analysis can provide vital characteristic information: sialic acid analysis. The degree of sialylation is typically related to the half-life of a glycoprotein in circulation because exposure of nonsialylated galactose residues leads to protein capture by asialoglycoprotein receptors and thus further catabolism.As indicated above, sialic acids form labile glycosidic linkages that can be cleaved using mild acid. This type of analysis is performed using a separate test sample because sialic acids would be destroyed by the time other monosaccharides are released with strong acid. Terminal sialic acids are also easily cleaved using sialidases.
Free sialic acids also can be analyzed in native form by HPAEC-PAD (using the appropriate elution conditions), or after fluorescent tagging (with DMB or OPD) followed by RP-HPLC with online fluorescence detection (22, 23). As above, both approaches can be validated to ensure accuracy, reproducibility, intermediate precision, robustness, and ruggedness. Furthermore, because analysis is based on the HPLC separation, it is possible to individually quantitate different sialic acids within one run (e.g., N-acetylneuraminic acid, Neu5Ac; N-glycolylneuraminic acid, Neu5Gc; their acetylated derivatives; and so on). If unknown fluorescent peaks are observed, the RP-HPLC method can be coupled with MS detection to identify the type of substituents present (24).
Sialic acid analysis has been a useful tool for evaluating the degree of variability in recombinant glycoproteins expressed by different cell lines relative to their human counterparts. It is also a valuable tool for evaluating process control because the expression of some species over others can result from process differences. Because sialic acid linkages are quite labile, monitoring the production of free sialic acid in a glycoprotein formulation is one way to assess product stability.
Other terminal monosaccharides can be assayed after exoglycosidase cleavage, followed by regular HPAEC-PAD or labeling and HPLC. The approach is particularly useful to evaluate the presence of terminal Gala(1–3) bound to an underlying galactose. That epitope elicits the antigenic reactions observed with xenotransplantation.
Oligosaccharide Profiling: Many analytical methods have been developed over the years for the analysis of oligosaccharide structures, just as with monosaccharides. The most widespread methods for oligosaccharide analysis involve cleavage of the chains from protein backbones, fractionation of the glycans produced, detection of each oligosaccharide, and establishment of a relative ratio for the signals obtained. It is also possible to directly analyze the mixture of glycans (native) by MALDI mass spectrometry (25).
Glycans can be analyzed in their native state (HPAEC-PAD) or after fluorescent tagging with a variety of reagents (e.g., 2-AA and 2-AB) (20, 21). Because only a few standard oligosaccharides species are commercially available — and those in small quantities only — the method is not usually validated to obtain an exact quantitation of oligosaccharide moieties against a known value, but rather to establish their relative ratio. This “profiling” strategy is a very powerful tool for monitoring consistency from lot to lot (the impact of process changes on the characteristics of a biotherapeutic product), and it is emerging as a key parameter for comparing brand-name and “generic” glycoproteins as follow-on biological products.
Oligosaccharides can be released from a protein backbone by chemical or enzymatic methods. The release should be quantitative, without glycan destruction or alteration. Chemical approaches include hydrazinolysis for release of N-and O-glycans (26), or carefully controlled reaction conditions to release only O-glycans (27), and ß-elimination that releases just O-glycans under specifically controlled conditions (28). Enzymatic approaches include PNGase F for release of N-glycans (29). Also, Endo H can be used for selective cleavage of high-mannose and most hybrid-type N-glycans (30)
. Chemical release of N- and O-glycans requires a higher level of expertise to perform, but it allows analysis of glycoproteins with unusual or unknown glycosylation without concern for missing structures because of selective cleavage.
Preparation for enzymatic deglycosylation usually entails reduction and alkylation of a glycoprotein and use of appropriate detergents. However, there is no enzyme appropriate for the quantitative release of all O-glycans. Thus, for analysis of N- and O-linked oligosaccharides, both chemical and enzymatic approaches need to be carefully set up using appropriate standards and performed reproducibly to assure reliable results. Unlike with monosaccharides, commercially available purified molecular standards for quantitation of oligosaccharide structures are limited and expensive. So it is typically impossible to empirically measure the accuracy of a technique against some known set of oligosaccharide values.
For oligosaccharide profiling, such limited quantities and species of standards curtail the use of HPAECPAD because the detector responses are different for each different species. So the actual quantities of those species can be different from how they appear. Nonetheless, this method provides exquisite separation of the released oligosaccharide species — based on their charge and structural features — as well as high sensitivity for underivatized glycans. Without derivatization, faster and simpler analysis are possible with fewer steps in which operational variability can be introduced. On the other hand, approaches involving fluorescent tagging show no differential responses to different glycans. However, sample derivatization and cleaning are required before analysis, and such manipulations also need standardization to provide reproducible results. Fluorescent-tagging methods can be coupled with MS when elution conditions are appropriate.
ABBREVIATIONS DEFINED
2-AA: 2-amino benzoic acid
2-AB: 2-amino-benzamide
CE: capillary electrophoresis
DMB: 1,2-diamino-4,5-methylendioxy-benzene
Endo-H: endo-beta-Nacetylglucosaminidase H
FID: flame ionization detector
GLC: gas–liquid chromatography
HPLC: high-performance liquid chromatography
HPAEC-PAD: high-pH, anion-exchange chromatography with pulsed amperometric detection
IEF: isoelectric focusing
IQ: installation qualification
LC: liquid chromatography
LIF: laser-induced fluorescence
MALDI: matrix assisted laser-desorption ionization
MW: molecular weight
MS: mass spectrometry
NMR: nuclear magnetic resonance
OPD: o-phenylendiamine
OQ: operational qualification
PQ: performance qualification
PNGase F: N-glycosidase F or endo-beta-N-Acetylglucosaminidase F peptide
QC: quality control
Q-Tof: quadrupole–time-of-flight
RP: reverse-phase
Monosaccharides
Fuc: fucose
Gal: galactose
GalNAc: N-Acetyl-galactosamine
GalUA: galacturonic acid
GlcNAc: N-Acetyl-glucosamine
GlcUA: glucuronic acid
Man: mannose
Sia: either N-Acetyl-neuraminic acid or sialic acid
Xyl: xylose
Both oligosaccharide profiling strategies require expertise and equipment that is properly set up and maintained. Ultimately, the choice of technology depends on the expertise and equipment available for successful method implementation.
Mixtures of oligosaccharides also can be analyzed using MALDI-MS to obtain their individual molecular weight (MW) information (25). The technique is unaffected by the presence of salts, detergents, and enzymes, so little clean-up is required before analysis. But desialylation does occur, making the method more suitable for studying nonsialylated species or oligosaccharide released and pretreated with neuraminidase.
Glycopeptide Mapping: A commonly used method for assessing reproducibility of protein expression and purification (as well as the identity and stability of purified glycoproteins) involves treatment with appropriate peptidases after reduction and alkylation, followed by RPHPLC analysis of the peptide fragments obtained. When a protein is glycosylated, the glycan-containing fragments are also observed, typically as broader peaks or clusters of unresolved peaks because of carbohydrate heterogeneity. Those are usually the peaks showing the most variability between lots and when conditions are changed, reflecting the sensibility of carbohydrate biosynthesis to small changes.
RP-HPLC methods are typically amenable to MS detection, so this approach provides a wealth of molecular information with little sample manipulation. There is no need for purification after enzymatic treatment, and artifact creation and loss of species present in small quantities are less likely than with some other methods. For glycan analysis it eliminates the selectivity associated with glycan release and reduces the number of manipulations and time required for analysis. It is often the first detailed analysis of glycosylation. The method is semiquantitative because different glycans can lead to different yields of ions in a spectrometer for different glycopeptides, and small, hydrophilic fragments might not be detected. But it provides an overall picture of the types of glycans present. Because in most cases peptidases can be chosen to target only one glycosylation site per peptide, this method provides an understanding of the differences between different glycosylation sites as well as the relative proportion of glycan species at each one. Of course, the technique must be tailored to each glycoprotein, but once established is reproducible and time-efficient.
Oligosaccharides can be quite large, and the difference between species is small, so oligopeptide mapping requires a high-end mass spectrometer (e.g., ion-trap, Q-TOF). LC-MS analysis can be a powerful tool for monitoring process control, establishing the impact of scaling up, or incorporating new steps during expression and purification. It can even help evaluate which specific glycosylation sites are the most susceptible to changes. This type of information is valuable for the overall understanding of the behavior of a specific glycoprotein, and it aids in designing protein stability studies and selecting formulations.
LC-MS oligosaccharide data can be used to validate an HPLC profiling strategy. After validation, QC release and stability test samples can be analyzed using the sample treatment and HPLC conditions, but monitored only by ultraviolet detection (omitting the MS part of the procedure). Chromatographic traces will provide unique “fingerprint profiles” that, when compared with a product reference standard analyzed concurrently with the sample, can serve as specifications for product identity, consistency, and in some cases stability.
Glycoform Profiling: Glycosylated proteins typically come in a variety of glycoforms, reflecting the fact that their carbohydrate moieties are diverse because of the lack of a “template” for their biosynthesis. Specific proteins are known for a characteristic set of glycoforms, each containing the same peptide backbone but differing in its attached oligosaccharides.
For sialylated glycoproteins, glycoforms have different mass-to-charge ratios because of the negative charge contribution of the sialic acid residues, so they can be easily separated by isoelectric focusing (IEF). IEF is routinely used to assess the distribution and abundance of glycoforms for biopharmaceutical products. IEF also can be valuable at early stages in selecting an expression system or cell culture conditions if an appropriate antibody is available for i
mmunostaining the protein of interest in the presence of all others. An aliquot of cell culture supernatant can be directly tested for glycoform expression. When quantitation of isoforms is desired, the gel IEF technique can be coupled to densitometric scanning. Both qualitative and quantitative IEF methods have been widely validated for use as QC release and stability-indicating assays for glycoproteins.
Recently, the developments in capillary electrophoresis have allowed good resolution of isoforms together with reliable quantitation. Some people feel that this method is on its way to replacing gel IEF. In many cases, a CE-IEF method can be validated using a well-characterized standard of a specific protein, thus providing a suitable assay for QC use.
Analysis of Intact Glycoproteins: Mass-spectrometric techniques can be used to evaluate the overall MW of intact glycoproteins. Each such protein requires the correct choice of MS technique. The MW of a glycoprotein such as erythropoietin can be accurately measured by MALDI-MS. However, in the case of an IgG, the heavy and light chains should be separated before analysis, so an LCMS approach is more suitable, with an instrument that provides accurate mass measurements.
Anomericity and Sequence Analysis of Complex Carbohydrates: In the context of biopharmaceutical production, when an expression system is well known and has been previously used, there is no actual need to confirm the anomericity or the exact linkage position for each monosaccharide constituent. These are defined by the biosynthetic machinery of the system in question.
Confirmation of anomericity (and sometimes linkage position) can be partially accomplished by examining the available exoglycosidases individually or in groups followed by reanalysis of the resulting oligosaccharide fragments with any of the methods described above. When using MALDI-MS, enzymatic treatments are directly accomplished on the MALDI target (25), an approach limited to the handful of enzymes commercially available.
Thorough evaluation of linkage types and positions requires complex chemistry approaches demanding a high level of training (e.g., in methylation analysis) and expensive instrumentation (e.g., NMR) (3). If sufficient quantities of purified glycans can be prepared, then nondestructive NMR analysis should be performed first, followed by whatever chemistry/MS approach is needed. Small quantities of oligosaccharides (2–5 nanomoles), even in mixtures, can yield good-quality 1H NMR data when spectrometers are fitted with sensitive probes. In some instances, modern analytical tools (MS approaches) have allowed identification of unexpected or unusual structural features (31, 33). With complex glycan heterogeneity (e.g., heavily O-glycosylated proteins), a combination of methods is required for complete characterization of these structures.
Anomericity and sequence analysis becomes relevant when exploring new expression systems, different organisms, and cell lines. A thorough understanding of their biosynthetic capabilities provides a guideline for expected structural motifs and their divergence from what is found in humans. Such understanding has led to well-known efforts in engineering enzymes (e.g., glycosyltransferases and glycosidases involved in human N-glycan biosynthesis) into promising systems for efficient protein expression (e.g., Pichia pastoris and Lemna). As new options are used, new analytical tools will become more common for ensuring that specific unwanted structural features are not present. Eventually, they will be part of the biotechnology tool box and ultimately might be converted into validatable, QC-amenable release and stability assays.
Knowledge Is PowerCarbohydrates are complex molecules containing a wealth of chemical information even in their simplest members, the monosaccharides. Although describing their structure can be an overwhelming task, a good understanding of their chemistry allows appropriate choice of analytical methods for each class of glycoconjugate. In addition, understanding their biochemistry helps us define the critical features that should be targeted for analysis depending on expression system or isolation method.
In the production of safe, efficacious, and consistent therapeutic glycoproteins, all critical features of complex carbohydrate structures can be assessed using current technology. Robust methods can be tailored to specific glycoproteins and even can be validated and used as release assays in controlled environments. Defining needs at each stage of development is key, both in the type of information and level of quantitation required. Although some methods require higher levels of expertise than others, they also offer more information. Often, more than one analytical approach is available for each purpose; the choice depends on a specific glycoprotein as well as existing instrumentation and expertise available. Expert advice can aid in initial method development and validation for a specific molecule. With suitably sound laboratory quality practices, a well-trained analyst should be able to perform these tests both reliably and reproducibly.
Analysis of glycosylation in glycoproteins has been covered here specifically, but similar principles and circumstances apply to the analysis of other glycoconjugates, even though the specific methods differ. Emerging technologies keep opening opportunities for more and better understanding. The more widespread use of powerful computers, sophisticated algorithms, and robotics will transform the current state-of-the-art approaches being tested by some experts into daily tools for biotech laboratories. And luckily for carbohydrate chemists, that means there are still many challenging (and rewarding) projects to work on.