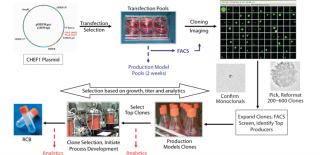
The standard cell line development pathway at many contract service organizations is based solely on selecting clones by growth and productivity with little or no analytical characterization of the expressed protein. The use of protein analytics to support selection of clones that match desired product quality is important for second-generation processes and will be of utmost importance for biosimilar development. CMC Biologics performs sophisticated protein characterization using state-of-the-art analytical equipment combined with our robust cell line development platform to generate and identify production-quality clones with desired product quality attributes in very rapid timeframes.
Implementing protein analysis into the cell line development process requires a robust platform capable of producing consistent results and timeframes. More importantly, it requires flexibility to enable matching of specific product quality attributes in the midst of the ongoing clone selection process. CMC Biologics’ CHEF1 expression platform is well suited to meet these stringent demands, as demonstrated in the following case study, wherein three separate antibody producing cell lines with specific glycan profiles were generated and identified in rapid timeframes.
Case Study
The CHEF1 expression platform includes three proprietary components — the CHEF1 plasmid; serum-free, suspension-adapted (SFSA) DG44 cells; and chemically defined CD-CIM1 media — optimized to work together in our standard cell line development platform to generate high-quality production clones in exceedingly rapid timeframes. A key feature of the rapid development timeline is that the CHEF1 plasmids promote high-level, stable expression of heterologous genes in Chinese hamster ovary (CHO) cell lines without gene amplification (1). CHEF1 plasmids employ upstream and downstream regulatory regions from the Chinese hamster elongation factor (CHEF) gene, a constitutively expressed housekeeping gene, to drive heterologous protein expression in hamster cells many fold better than the human EF promoter or the viral CMVpromoter.
The selection strategy for cloning CHEF1 cell lines has been optimized using our SFSA cells, which have been adapted to grow in CD-CIM1 media. The entire development process, including cloning, is performed in the absence of any animal-derived products.
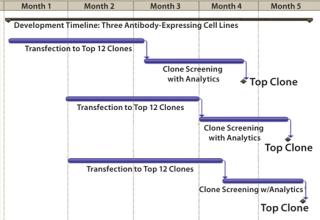
Three different monoclonal antibody (MAb) expressing cell lines were generated using the standard CHEF1 platform. Beginning with transfection, selection of a production clone suitable for bioreactor process development based on growth and titer alone would typically require 12 weeks (Figure 1). In this particular case study, the platform was modified to include early stage analytics for the top 12 clones selected by growth and titer, then further advancement of the top 4 clones matching the desired product quality profile into bioreactors, producing larger quantities of protein for supplemental analysis. The selection of single clones with desirable early stage productivity and matching specific protein characteristics of the control antibodies (each made by an independent manufacturer) was obtained in 16–17 weeks from transfection (Figure 2).
Transfection pools were expanded to provide early stage material for purification and analytical development. The titers for the three transfection pools, generated in five to six weeks, ranged from 400 to 600 mg/Land were sufficient to initiate methods development. Cells from the transfection pools were seeded into 96-well plates by limiting dilution, grown for two weeks, and then screened by a high-throughput FACS-based assay to identify approximately 24–48 clones with the highest titers. Monoclonality was determined by imaging colony outgrowth in cloning plates with the CloneSelect imager (Genetix Ltd., UK), identifying wells with single isolated colonies, then confirming the colony derived from a single cell (Figure 1) in the Day 1 images.
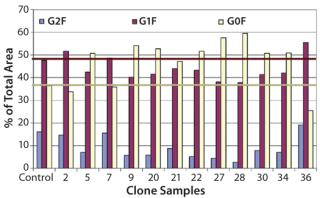
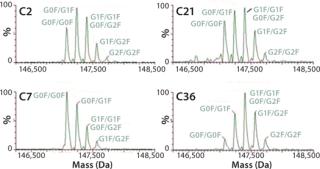
The selected clones were further consolidated by titer determination and run in fed-batch shake flask and bioreactor production models to produce protein for early stage analytics. The battery of analytical assays performed included reduced and nonreduced SDS-PAGE; size-exclusion (SE) HPLC; ion-exchange (IE) HPLC; glycan analysis by IE-HPLC on the liberated, fluoresceinated sugars; and intact mass analysis on the native antibody. Most of these assays indicated that the MAbs produced with CHEF1 expression system were similar to the control protein and each other. Subtle SE-HPLC differences were detected, whereas the majority of clones produced >94% correctly sized protein with some higher MW species making up the difference. The real differences were seen in the glycosylation, as determined by both glycan and intact mass analysis. Figure 3 shows an example of the galactosylation (G0, G1, and G2) differences seen in the MAb1 clones in comparison to the control MAb. The top four clones for this MAb were further analyzed by intact mass analysis using the Synapt mass spectrometer (Waters Corp., USA), showing exquisitely fine resolution of the whole antibody protein molecule, yielding more information about glycosylation and other posttranslational modifications (Figure 4).
<
p>
In summary, the top clones isolated from each of the three MAb cell line development projects have competitive early stage productivities (0.8–1.0 g/L) and very similar protein characteristics to the control proteins we set out to match. We envision that these combined capabilities place CMC Biologics in a unique position to offer comprehensive services to clients with more demanding cell line development projects and complement our expertise in later-stage process optimization.
Author Details
Howard Clarke, PhD, is director of upstream process development at CMC Biologics, 22021 20th Avenue SE, Bothell, WA 98021; 1-425-485-1900, fax 1-425-486-0300; www.cmcbio.com.