Cell counting and viability assessment are routine but essential parts of bioprocessing procedures, and accuracy, reproducibility, and standardization are essential to successful outcomes. Traditional methods produce variable results because they rely on subjective discrimination of dye-stained cells using a hemacytometer and microscope. Substantial operator-to-operator variability in the cell counts is common, even when one researcher examines the same sample twice.
Guava technologies offers a solution: a simple, accurate, and reproducible absolute cell counting and viability assay that overcomes many of the drawbacks of the manual method. The simple and robust Guava ViaCount® assay eliminates statistical variation by sampling large numbers of single cells, and it removes operator variability by categorizing cells as live, dead, or apoptotic by quantifying fluorescence staining.
Accurate Cell Counting via MicrocytometryThe Guava ViaCount assay runs on all of the compact, benchtop Guava Systems: PCA™, PCA-96, EasyCyte™, and EasyCyte Mini. These systems use proprietary microcytometry technology with fluorescence-based cellular analysis. Guava’s unique integration and optimization of instruments, software, and reagents offers
-
Easy absolute cell counting
-
Analysis of hundreds of cells per second
-
A wide range of biological assays on a single platform
-
Automatic processing of dozens of samples per run when using the PCA-96 and EasyCyte systems.
Based on a dual fluorescent staining reagent, Guava ViaCount assay relies on membrane exclusion of a fluorescent dye and a nuclear stain to distinguish live cells from debris, dead cells, and mid-late stage apoptotic cells. Users set markers to determine how cells will be identified based on each cell’s fluorescence intensity for the nuclear and dead cell dyes. Guava CytoSoft™ software and ViaCount assay together offer a complete set of adjustable thresholds and settings for optimization across a wide range of cell types.
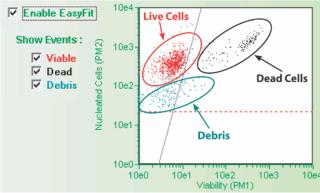
Cell Counting By Standardized Pattern RecognitionAn enhanced version of the Guava ViaCount assay incorporates the Guava EasyFit™ feature, which offers even greater automation and standardization for viability and cell counting. A powerful computer-assisted population analysis tool, the EasyFit feature implements a unique clustering algorithm that interprets cellular distribution patterns in a 3-D matrix; groups of cells are defined and counted using three characteristics of each cell, including the fluorescence intensity of staining with the live and dead cell dyes. Individual events are automatically separated based on their labeling patterns and color coded according to whether they are live, dead, or noncellular debris.
The EasyFit feature streamlines cell counting and distribution analysis by removing human analysis. Variability in setting thresholds or defining groups of cells is eliminated because standardized counting methods are applied providing more consistent quality data, even from novice users.
Debris IndexTypically, thresholds are set manually prior to data acquisition to exclude debris from being counted in cell viability calculations, so resulting data show only information about live, dead, and apoptotic fractions. To quantify the amount of cellular debris, the Guava EasyFit feature detects cellular debris and reports a value called the Debris Index, which represents the percentage of total events that are categorized as debris. The debris index can be a valuable indicator for monitoring bioreactor performance, where tracking increases in debris can be helpful in understanding if or when to modify bioreactor conditions.
SummaryThe ViaCount EasyFit feature from Guava Technologies improves the ease of use of the Guava Systems and provides increased functionality for characterization of debris within cell samples. Once validated for cell lines of interest, enabling the EasyFit feature can improve workflow, reduce training time, and standardize analysis methods from user to user. By analyzing cells in three dimensions and evaluating how populations of cells differentiate into clusters, The EasyFit feature can more accurately differentiate distribution patterns and improve data quality.
Guava and ViaCount are registered trademarks, and CytoSoft, EasyFit, PCA, and EasyCyte are trademarks of Guava technologies Inc. © 2006 Guava technologies Inc. All rights reserved.